Characterizing the dynamics of proteasome complexes by proteomics approaches - PubMed (original) (raw)
Review
. 2014 Dec 10;21(17):2444-56.
doi: 10.1089/ars.2013.5815. Epub 2014 Mar 27.
Affiliations
- PMID: 24423446
- PMCID: PMC4241863
- DOI: 10.1089/ars.2013.5815
Review
Characterizing the dynamics of proteasome complexes by proteomics approaches
Robyn M Kaake et al. Antioxid Redox Signal. 2014.
Abstract
Significance: The proteasome is the degradation machine of the ubiquitin-proteasome system, which is critical in controlling many essential biological processes. Aberrant regulation of proteasome-dependent protein degradation can lead to various human diseases, and general proteasome inhibitors have shown efficacy for cancer treatments. Though clinically effective, current proteasome inhibitors have detrimental side effects and, thus, better therapeutic strategies targeting proteasomes are needed. Therefore, a comprehensive characterization of proteasome complexes will provide the molecular details that are essential for developing new and improved drugs.
Recent advances: New mass spectrometry (MS)-based proteomics approaches have been developed to study protein interaction networks and structural topologies of proteasome complexes. The results have helped define the dynamic proteomes of proteasome complexes, thus providing new insights into the mechanisms underlying proteasome function and regulation.
Critical issues: The proteasome exists as heterogeneous populations in tissues/cells, and its proteome is highly dynamic and complex. In addition, proteasome complexes are regulated by various mechanisms under different physiological conditions. Consequently, complete proteomic profiling of proteasome complexes remains a major challenge for the field.
Future directions: We expect that proteomic methodologies enabling full characterization of proteasome complexes will continue to evolve. Further advances in MS instrumentation and protein separation techniques will be needed to facilitate the detailed proteomic analysis of low-abundance components and subpopulations of proteasome complexes. The results will help us understand proteasome biology as well as provide new therapeutic targets for disease diagnostics and treatment.
Figures
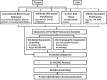
**FIG. 1.
Defining proteasome dynamics to understand proteasome biology. PTMs, posttranslational modifications.
**FIG. 2.
The general workflow for isolating and analyzing proteasome complexes. DEAE, diethylaminoethyl; HB, histidine-biotin; IEF, isoelectric focusing; LC-MS/MS, liquid chromatography tandem mass spectrometry; SDS-PAGE, sodium dodecyl sulfate polyacrylamide gel electrophoresis; TAP, tandem affinity purification; UBL, ubiquitin-like.
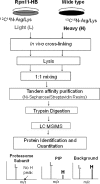
**FIG. 3.
Schematic diagram of the QTAX strategy. Cells expressing a His-Bio-tagged proteasome subunit (i.e., Rpn11-HB) and a wild-type strain were grown in a light (12C14N-Arg/Lys) and a heavy (13C15N-Arg/Lys) containing media, respectively. After in vivo formaldehyde cross-linking, equal amounts of cell lysates were mixed for HB-tag based tandem affinity purification. The bound proteins were on-bead digested and analyzed by LC-MS/MS. Specific PIPs can be differentiated from background proteins with their SILAC ratios (L/H), based on the relative abundance ratios of Arg/Lys containing peptide pairs. Three groups of proteins were generally identified: (i) proteasome subunits; (ii) PIPs; and (iii) background proteins. PIPs, proteasome interacting proteins; QTAX, quantitative analysis of tandem affinity purified in vivo cross-linked (x) protein complexes.
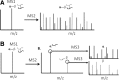
**FIG. 4.
MS sequencing of inter-linked peptides. (A) MS2 analysis of an noncleavable inter-linked peptide (α-β) results in a complex spectrum containing sequence ions from both peptides, which prevents it from being searched effectively by conventional database search engines; (B) MS2 analysis of an MS-cleavable inter-linked peptide detected in MS1, for example, a DSSO cross-linked peptide, results in the physical separation of α and β peptides, which enables their subsequent MS3 sequencing for unambiguous identification using conventional database search engines. DSSO, disuccinimidyl sulfoxide; MS, mass spectrometry.
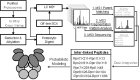
**FIG. 5.
The general workflow of DSSO-based XL-MS strategy for structure modeling of proteasome complexes. The purified proteasome complex was cross-linked in vitro with DSSO; the resulting products were digested, and subsequently separated and analyzed by LC MSn. The resulting data provide three lines of evidence supporting the identification of DSSO cross-linked peptides: (i) Potential cross-linked peptides are determined based on the parent mass measured in MS1 through database searching using the MS-Bridge tool in Protein Prospector; (ii) characteristic fragmentation patterns from low-energy cleavage of DSSO cross-linked peptides results in a simple MS2 spectrum that contain peaks with specific mass relationships to their parent mass; and (iii) MS3 analysis of the individual peptide fragment ions detected in MS2 provides unambiguous peptide identification using conventional database searching methods such as Batch-Tag in Protein Prospector. Together, these three pieces of information are integrated and analyzed to enable the confident identification of cross-linked peptides. Cross-links that have been identified for the yeast 19S RP, representing binary inter-subunit interactions, are then used in a probabilistic modeling analysis to derive the spatial ordering of the 19S subcomplexes with the highest probability. MSn, multistage tandem mass spectrometry; RP, regulatory particle; XL-MS, cross-linking mass spectrometry.
Similar articles
- Proteomics to study the diversity and dynamics of proteasome complexes: from fundamentals to the clinic.
Bousquet-Dubouch MP, Fabre B, Monsarrat B, Burlet-Schiltz O. Bousquet-Dubouch MP, et al. Expert Rev Proteomics. 2011 Aug;8(4):459-81. doi: 10.1586/epr.11.41. Expert Rev Proteomics. 2011. PMID: 21819302 Review. - Exploring proteasome complexes by proteomic approaches.
Drews O, Zong C, Ping P. Drews O, et al. Proteomics. 2007 Apr;7(7):1047-58. doi: 10.1002/pmic.200600574. Proteomics. 2007. PMID: 17390294 Review. - Proteomics of proteasome complexes and ubiquitinated proteins.
Wang X, Guerrero C, Kaiser P, Huang L. Wang X, et al. Expert Rev Proteomics. 2007 Oct;4(5):649-65. doi: 10.1586/14789450.4.5.649. Expert Rev Proteomics. 2007. PMID: 17941820 Review. - Subcellular distribution and dynamics of active proteasome complexes unraveled by a workflow combining in vivo complex cross-linking and quantitative proteomics.
Fabre B, Lambour T, Delobel J, Amalric F, Monsarrat B, Burlet-Schiltz O, Bousquet-Dubouch MP. Fabre B, et al. Mol Cell Proteomics. 2013 Mar;12(3):687-99. doi: 10.1074/mcp.M112.023317. Epub 2012 Dec 13. Mol Cell Proteomics. 2013. PMID: 23242550 Free PMC article. - Molecular Details Underlying Dynamic Structures and Regulation of the Human 26S Proteasome.
Wang X, Cimermancic P, Yu C, Schweitzer A, Chopra N, Engel JL, Greenberg C, Huszagh AS, Beck F, Sakata E, Yang Y, Novitsky EJ, Leitner A, Nanni P, Kahraman A, Guo X, Dixon JE, Rychnovsky SD, Aebersold R, Baumeister W, Sali A, Huang L. Wang X, et al. Mol Cell Proteomics. 2017 May;16(5):840-854. doi: 10.1074/mcp.M116.065326. Epub 2017 Mar 14. Mol Cell Proteomics. 2017. PMID: 28292943 Free PMC article.
Cited by
- Neuromodulatory effect of vardenafil on aluminium chloride/D-galactose induced Alzheimer's disease in rats: emphasis on amyloid-beta, p-tau, PI3K/Akt/p53 pathway, endoplasmic reticulum stress, and cellular senescence.
Awad HH, Desouky MA, Zidan A, Bassem M, Qasem A, Farouk M, AlDeab H, Fouad M, Hany C, Basem N, Nader R, Alkalleny A, Reda V, George MY. Awad HH, et al. Inflammopharmacology. 2023 Oct;31(5):2653-2673. doi: 10.1007/s10787-023-01287-w. Epub 2023 Jul 17. Inflammopharmacology. 2023. PMID: 37460908 Free PMC article. - Proteomic analysis of affinity-purified extracellular proteasomes reveals exclusively 20S complexes.
Kulichkova VA, Artamonova TO, Lyublinskaya OG, Khodorkovskii MA, Tomilin AN, Tsimokha AS. Kulichkova VA, et al. Oncotarget. 2017 Nov 1;8(60):102134-102149. doi: 10.18632/oncotarget.22230. eCollection 2017 Nov 24. Oncotarget. 2017. PMID: 29254231 Free PMC article. - Proteasome Dysfunction Associated to Oxidative Stress and Proteotoxicity in Adipocytes Compromises Insulin Sensitivity in Human Obesity.
Díaz-Ruiz A, Guzmán-Ruiz R, Moreno NR, García-Rios A, Delgado-Casado N, Membrives A, Túnez I, El Bekay R, Fernández-Real JM, Tovar S, Diéguez C, Tinahones FJ, Vázquez-Martínez R, López-Miranda J, Malagón MM. Díaz-Ruiz A, et al. Antioxid Redox Signal. 2015 Sep 1;23(7):597-612. doi: 10.1089/ars.2014.5939. Epub 2015 Mar 31. Antioxid Redox Signal. 2015. PMID: 25714483 Free PMC article. - Deciphering preferential interactions within supramolecular protein complexes: the proteasome case.
Fabre B, Lambour T, Garrigues L, Amalric F, Vigneron N, Menneteau T, Stella A, Monsarrat B, Van den Eynde B, Burlet-Schiltz O, Bousquet-Dubouch MP. Fabre B, et al. Mol Syst Biol. 2015 Jan 5;11(1):771. doi: 10.15252/msb.20145497. Mol Syst Biol. 2015. PMID: 25561571 Free PMC article. - Targeting the ubiquitin-proteasome system in heart disease: the basis for new therapeutic strategies.
Drews O, Taegtmeyer H. Drews O, et al. Antioxid Redox Signal. 2014 Dec 10;21(17):2322-43. doi: 10.1089/ars.2013.5823. Epub 2014 Oct 1. Antioxid Redox Signal. 2014. PMID: 25133688 Free PMC article. Review.
References
- Ang XL. and Wade Harper J. SCF-mediated protein degradation and cell cycle control. Oncogene 24: 2860–2870, 2005 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R21 CA161807/CA/NCI NIH HHS/United States
- R01GM074830/GM/NIGMS NIH HHS/United States
- R01 GM106003/GM/NIGMS NIH HHS/United States
- R01 GM074830/GM/NIGMS NIH HHS/United States
- R01GM106003/GM/NIGMS NIH HHS/United States
- R21CA161807/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources