The Drosophila insulin-degrading enzyme restricts growth by modulating the PI3K pathway in a cell-autonomous manner - PubMed (original) (raw)
The Drosophila insulin-degrading enzyme restricts growth by modulating the PI3K pathway in a cell-autonomous manner
Diego Galagovsky et al. Mol Biol Cell. 2014 Mar.
Abstract
Mammalian insulin-degrading enzyme (IDE) cleaves insulin, among other peptidic substrates, but its function in insulin signaling is elusive. We use the Drosophila system to define the function of IDE in the regulation of growth and metabolism. We find that either loss or gain of function of Drosophila IDE (dIDE) can restrict growth in a cell-autonomous manner by affecting both cell size and cell number. dIDE can modulate Drosophila insulin-like peptide 2 levels, thereby restricting activation of the phosphatidylinositol-3-phosphate kinase pathway and promoting activation of Drosophila forkhead box, subgroup O transcription factor. Larvae reared in high sucrose exhibit delayed developmental timing due to insulin resistance. We find that dIDE loss of function exacerbates this phenotype and that mutants display increased levels of circulating sugar, along with augmented expression of a lipid biosynthesis marker. We propose that dIDE is a modulator of insulin signaling and that its loss of function favors insulin resistance, a hallmark of diabetes mellitus type II.
Figures
FIGURE 1:
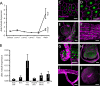
dIDE gene expression pattern. (A) Temporal variation of the expression of dide throughout the life cycle, as determined by qRT-PCR. Error bars represent SD (three independent experiments). (B) Expression of dide in third-instar larval organs. mRNA levels were determined by qRT-PCR. Error bars represent SD (three independent experiments). (C–J) Expression of dide in third-instar larval organs, as revealed by expression of a nuclear GFP reporter under the control of the dide promoter; representative organs. (C) Salivary glands, (D) fat body, (E) anterior gut and proventriculus, (F) midgut, (G) hindgut, (H) wing imaginal disc, (I) tracheae, and (J) brain. Bars, 50 μm.
FIGURE 2:
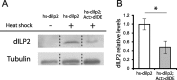
dIDE expression reduces dILP2 levels. (A) Anti-dILP2 Western blot of whole third- instar larval extracts prepared from individuals overexpressing dILP2 and coexpressing or not dIDE. Expression of dIDE provokes reduction of dILP2 levels. (B) Quantification of the Western blot. Error bars represent SEM (*p < 0.05; Student's t test, n = 5).
FIGURE 3:
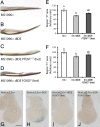
dIDE provokes growth reduction in a cell-autonomous manner. (A) Pupae from individuals that express transgenic dIDE ubiquitously are significantly smaller than those of control siblings, as determined by their pupal volume (see Materials and Methods). Error bars represent SEM (**p < 0.001; Student's t test; n ≥ 30 in three independent experiments). (B, C) Expression of dIDE in the dorsal compartment of the wing imaginal disc generates upwardly curved wings, indicating reduction of growth of this compartment. (D, E) Expression of dIDE at the posterior compartment of the disc provokes a reduction of the posterior half of the wing; this was estimated by measuring the area of the wing region D (indicated by the broken line in D, E). (F) Quantification of wing areas marked in D and E. Error bars represent SEM (**p < 0.001; Student's t test; n ≥ 20 in three independent experiments). (G) Wing hair density at the posterior compartment of the wing. Following expression of dIDE, hair density increased, indicating that cell size was reduced. Cell size reduction accounts only partially for reduction of the area of the wing posterior compartment; compare delta values in F (19.1%) and G (11.2%). The remaining reduction of the area is due to decreased number of cells in the compartment. Error bars represent SEM (**p < 0.001; Student's t test; n ≥ 20 in three independent experiments).
FIGURE 4:
Reduction of growth upon dIDE expression is mediated by the PI3K pathway. (A–D) The wing curvature provoked by the expression dIDE at the dorsal compartment of the imaginal disc (A, B) is partially suppressed in flies lacking one dose of PTEN (C) or FOXO (D). (E, F) Reduction of the area of the posterior compartment provoked by the expression of dIDE under control of an _en_-Gal4 driver (assessed by determination of the wing region D) is partially reverted in individuals lacking one dose of PTEN (E) or FOXO (F). These results (A–F) indicate that the PI3K pathway mediates dIDE-dependent growth reduction through the regulation of FOXO. Error bars represent SEM (p < 0.05; one-way analysis of variance (ANOVA) with Tukey post hoc test; means with a letter in common are not significantly different; n ≥ 20 in three independent experiments). (G, H) The FOXO target gene thor is induced at the posterior compartment of the wing imaginal disc upon expression of dIDE in the same compartment, suggesting that dIDE-dependent inactivation of the PI3K pathway and activation of FOXO is cell autonomous. In foxo25 (I) or foxo21 (J) heterozygous mutant larvae, dIDE-dependent thor transcription is reduced, confirming that FOXO mediates thor-LacZ induction in this setting. Bar, 100 μm.
FIGURE 5:
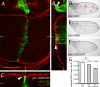
dIDE localizes in the cytoplasm and plasma membrane of wing imaginal disc cells. Expression of a dIDE:GFP fusion protein was induced in the wing imaginal disc through a _ptc_-Gal4 driver. (A) The fusion protein can be detected only in the narrow stripe of cells (two to four cells) that expresses patched, without any indication that the protein can move away into neighboring cells. (B) Longitudinal and (C) transverse _z_-sections of the wing disc showing that the fusion protein accumulates in the cytoplasm and plasma membrane (arrowheads) of _ptc_-expressing cells; scale bar, 50 μm. (D–F) The expression of dIDE (E) or dIDE:GFP (F) under control of the _ptc_-Gal4 driver reduces the distance between veins III and IV (shown in D) as compared with control wings expressing GFP alone (D). (G) Quantification of the effect of dIDE or dIDE:GFP expression on the reduction of the distance between veins III and IV. The effect of dIDE:GFP (30% reduction) is stronger than that of dIDE alone (12.6% reduction). The distance between veins III and IV is indicated as a (green line) in D. The distance a was normalized to the length of vein III, indicated as b (red line; **p < 0.001; one-way ANOVA with Tukey post hoc test; n ≥ 20 in three independent experiments).
FIGURE 6:
dIDE loss of function provokes reduction of growth and sensitization to a high-sugar diet. (A) dIDEKO pupae are significantly smaller than those of control (WT) siblings, as assessed by calculating their pupal volume (see Materials and Methods). Error bars represent SEM (**p < 0.001; Student's t test; n ≥ 30 in three independent experiments). (B, C) The dorsal compartment of the wing imaginal disc is autonomously reduced upon dIDE RNAi expression. Disc compartments can be visualized by the expression of Wingless (Wg, green); d, disc dorsal compartment; v, disc ventral compartment. Bar, 100 μm. (D, E) dIDERNAi-dependent autonomous growth reduction of the dorsal compartment resulted in wings curved upward. (F, G) dIDERNAi expression at the posterior compartment of the wing disc provoked autonomous reduction of this compartment, as assessed by determination of the area of region D. Expression of RNAi in PTEN (F) or FOXO (G) heterozygous mutants caused strong suppression of growth impairment. Error bars represent SEM (p < 0.05; one way ANOVA with Tukey post hoc test; means with a letter in common are not significantly different; n ≥ 20 in three independent experiments). (H) dIDEKO and control larvae pupariate almost at the same time in standard culture medium. In a high-sugar culture medium, significant delay of pupariation of wild-type individuals occurred; this delay was increased in dIDEKO larvae. Error bars represent SD (**p < 0.001; two-way ANOVA with Duncan post hoc test; n = 60 in six independent experiments). (I) In WT adult flies, trehalose levels in hemolymph dropped significantly after 24-h starvation; in dIDEKO individuals subjected to starvation, trehalose levels remained similar to those of well-fed animals. Error bars represent SEM (ns, non significant; *p < 0.05; two-way ANOVA with Tukey post hoc test; n = 5). (J) dIDEKO flies display augmented levels of the acetyl CoA carboxylase (ACC) transcript, used as an indicator of TAG metabolism. In high-sugar medium, ACC expression increased significantly in wild-type larvae, but ACC transcript levels in dIDEKO individuals were even higher. Error bars represent SEM (**p < 0.001; two-way ANOVA with Tukey post hoc test; n = 3).
Similar articles
- The Hippo pathway integrates PI3K-Akt signals with mechanical and polarity cues to control tissue growth.
Borreguero-Muñoz N, Fletcher GC, Aguilar-Aragon M, Elbediwy A, Vincent-Mistiaen ZI, Thompson BJ. Borreguero-Muñoz N, et al. PLoS Biol. 2019 Oct 15;17(10):e3000509. doi: 10.1371/journal.pbio.3000509. eCollection 2019 Oct. PLoS Biol. 2019. PMID: 31613895 Free PMC article. - Cloning and expression of the cDNA for a Drosophila insulin-degrading enzyme.
Kuo WL, Gehm BD, Rosner MR. Kuo WL, et al. Mol Endocrinol. 1990 Oct;4(10):1580-91. doi: 10.1210/mend-4-10-1580. Mol Endocrinol. 1990. PMID: 2126597 - Insulin-degrading enzyme antagonizes insulin-dependent tissue growth and Abeta-induced neurotoxicity in Drosophila.
Tsuda M, Kobayashi T, Matsuo T, Aigaki T. Tsuda M, et al. FEBS Lett. 2010 Jul 2;584(13):2916-20. doi: 10.1016/j.febslet.2010.05.010. Epub 2010 May 21. FEBS Lett. 2010. PMID: 20493190 - Protocols to Study Growth and Metabolism in Drosophila.
Strassburger K, Teleman AA. Strassburger K, et al. Methods Mol Biol. 2016;1478:279-290. doi: 10.1007/978-1-4939-6371-3_17. Methods Mol Biol. 2016. PMID: 27730589 Review. - Diabetic larvae and obese flies-emerging studies of metabolism in Drosophila.
Baker KD, Thummel CS. Baker KD, et al. Cell Metab. 2007 Oct;6(4):257-66. doi: 10.1016/j.cmet.2007.09.002. Cell Metab. 2007. PMID: 17908555 Free PMC article. Review.
Cited by
- Role of Serotonin Transporter in Eye Development of Drosophila melanogaster.
Pham TLA, Binh TD, Liu G, Nguyen TQC, Nguyen YDH, Sahashi R, Men TT, Kamei K. Pham TLA, et al. Int J Mol Sci. 2020 Jun 8;21(11):4086. doi: 10.3390/ijms21114086. Int J Mol Sci. 2020. PMID: 32521639 Free PMC article. - Ensemble cryoEM elucidates the mechanism of insulin capture and degradation by human insulin degrading enzyme.
Zhang Z, Liang WG, Bailey LJ, Tan YZ, Wei H, Wang A, Farcasanu M, Woods VA, McCord LA, Lee D, Shang W, Deprez-Poulain R, Deprez B, Liu DR, Koide A, Koide S, Kossiakoff AA, Li S, Carragher B, Potter CS, Tang WJ. Zhang Z, et al. Elife. 2018 Mar 29;7:e33572. doi: 10.7554/eLife.33572. Elife. 2018. PMID: 29596046 Free PMC article. - Bone marrow mesenchymal stem cell-derived exosomes reduce insulin resistance and obesity in mice via the PI3K/AKT signaling pathway.
Shi H, Hao X, Sun Y, Zhang H, Zhao Y, Wang B, Lu J, Hou W, Yan Y, Yu X, Xue L, Luo X, Wang H. Shi H, et al. FEBS Open Bio. 2023 Jun;13(6):1015-1026. doi: 10.1002/2211-5463.13615. Epub 2023 May 2. FEBS Open Bio. 2023. PMID: 37073893 Free PMC article. - Inhibition of Insulin Degrading Enzyme to Control Diabetes Mellitus and its Applications on some Other Chronic Disease: a Critical Review.
Azam MS, Wahiduzzaman M, Reyad-Ul-Ferdous M, Islam MN, Roy M. Azam MS, et al. Pharm Res. 2022 Apr;39(4):611-629. doi: 10.1007/s11095-022-03237-7. Epub 2022 Apr 4. Pharm Res. 2022. PMID: 35378698 Review. - Fucoidan alleviates high sucrose-induced metabolic disorders and enhances intestinal homeostasis through modulation of Notch signaling.
Liu J, Xia W, Wu Q, Zhang Y, Wu Y, Li B, Chen F, Du X, Wu S, Yang Y, Gao Y, Wu M, Su L, Tong H. Liu J, et al. J Adv Res. 2025 May;71:189-207. doi: 10.1016/j.jare.2024.05.034. Epub 2024 May 31. J Adv Res. 2025. PMID: 38825316 Free PMC article.
References
- Authier F, Rachubinski RA, Posner BI, Bergeron JJ. Endosomal proteolysis of insulin by an acidic thiol metalloprotease unrelated to insulin degrading enzyme. J Biol Chem. 1994;269:3010–3016. - PubMed
- Backer JM, Kahn CR, White MF. The dissociation and degradation of internalized insulin occur in the endosomes of rat hepatoma cells. J Biol Chem. 1990;265:14828–14835. - PubMed
- Barolo S, Carver LA, Posakony JW. GFP and beta-galactosidase transformation vectors for promoter/enhancer analysis in Drosophila. BioTechniques. 2000;29:726, 728, 730, 732. - PubMed
- Birdsall K, Zimmerman E, Teeter K, Gibson G. Genetic variation for the positioning of wing veins in Drosophila melanogaster. Evol Dev. 2000;2:16–24. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases