Toxin-antitoxin systems as multilevel interaction systems - PubMed (original) (raw)
Review
Toxin-antitoxin systems as multilevel interaction systems
Nathalie Goeders et al. Toxins (Basel). 2014.
Abstract
Toxin-antitoxin (TA) systems are small genetic modules usually composed of a toxin and an antitoxin counteracting the activity of the toxic protein. These systems are widely spread in bacterial and archaeal genomes. TA systems have been assigned many functions, ranging from persistence to DNA stabilization or protection against mobile genetic elements. They are classified in five types, depending on the nature and mode of action of the antitoxin. In type I and III, antitoxins are RNAs that either inhibit the synthesis of the toxin or sequester it. In type II, IV and V, antitoxins are proteins that either sequester, counterbalance toxin activity or inhibit toxin synthesis. In addition to these interactions between the antitoxin and toxin components (RNA-RNA, protein-protein, RNA-protein), TA systems interact with a variety of cellular factors, e.g., toxins target essential cellular components, antitoxins are degraded by RNAses or ATP-dependent proteases. Hence, TA systems have the capacity to interact with each other at different levels. In this review, we will discuss the different interactions in which TA systems are involved and their implications in TA system functions and evolution.
Figures
Figure 1
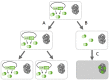
The addiction phenomenon. Toxin-antitoxin (TA) systems participate in plasmid maintenance in growing bacterial populations by a mechanism called addiction or post-segregational killing. Addiction relies on the differential stability of the toxin and antitoxin. A: Daughter-bacteria that inherit a plasmid copy encoding the ccd (
c
ontrol of
c
ell
d
eath) toxin-antitoxin system grow normally. B: Daughter-bacteria that do not inherit a plasmid copy still have antitoxin-toxin complexes in their cytoplasm. C: The CcdA antitoxin (light green) is degraded by the Lon protease, while the CcdB toxin (dark green) is stable. CcdB is, therefore, liberated from the CcdA-CcdB complex and is able to interact with DNA-gyrase, a class II topoisomerase. The interaction of CcdB with DNA-gyrase inhibits DNA replication and leads eventually to cell death. Addiction leads to the selective killing of plasmid-free daughter bacteria and increases plasmid prevalence in the bacterial population.
Figure 2
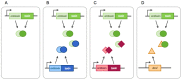
Interactions between TA systems. (A) In most cases, antitoxins and toxins only interact with their cognate partners; (B) chromosomal systems can interact with homologous systems, such as for anti-addiction; (C) TA systems can bind non-canonical antitoxins and toxins, which can lead to network formation and (D) a toxin can be inhibited by a protein not related to TA systems, as in the case of Dmd of the T4 phage.
Similar articles
- Type II Toxin-Antitoxin Systems in Pseudomonas aeruginosa.
Li M, Guo N, Song G, Huang Y, Wang L, Zhang Y, Wang T. Li M, et al. Toxins (Basel). 2023 Feb 17;15(2):164. doi: 10.3390/toxins15020164. Toxins (Basel). 2023. PMID: 36828478 Free PMC article. Review. - Regulating toxin-antitoxin expression: controlled detonation of intracellular molecular timebombs.
Hayes F, Kędzierska B. Hayes F, et al. Toxins (Basel). 2014 Jan 15;6(1):337-58. doi: 10.3390/toxins6010337. Toxins (Basel). 2014. PMID: 24434949 Free PMC article. Review. - Structure-function analysis of VapB4 antitoxin identifies critical features of a minimal VapC4 toxin-binding module.
Jin G, Pavelka MS Jr, Butler JS. Jin G, et al. J Bacteriol. 2015 Apr;197(7):1197-207. doi: 10.1128/JB.02508-14. Epub 2015 Jan 26. J Bacteriol. 2015. PMID: 25622615 Free PMC article. - Structural Determinants for Antitoxin Identity and Insulation of Cross Talk between Homologous Toxin-Antitoxin Systems.
Walling LR, Butler JS. Walling LR, et al. J Bacteriol. 2016 Nov 18;198(24):3287-3295. doi: 10.1128/JB.00529-16. Print 2016 Dec 15. J Bacteriol. 2016. PMID: 27672196 Free PMC article. - Toxin-Antitoxin Systems of Staphylococcus aureus.
Schuster CF, Bertram R. Schuster CF, et al. Toxins (Basel). 2016 May 5;8(5):140. doi: 10.3390/toxins8050140. Toxins (Basel). 2016. PMID: 27164142 Free PMC article. Review.
Cited by
- Multitasking SecB chaperones in bacteria.
Sala A, Bordes P, Genevaux P. Sala A, et al. Front Microbiol. 2014 Dec 5;5:666. doi: 10.3389/fmicb.2014.00666. eCollection 2014. Front Microbiol. 2014. PMID: 25538690 Free PMC article. Review. - Characterization of DinJ-YafQ toxin-antitoxin module in Tetragenococcus halophilus: activity, interplay, and evolution.
Luo X, Lin J, Yan J, Kuang X, Su H, Lin W, Luo L. Luo X, et al. Appl Microbiol Biotechnol. 2021 May;105(9):3659-3672. doi: 10.1007/s00253-021-11297-9. Epub 2021 Apr 20. Appl Microbiol Biotechnol. 2021. PMID: 33877415 - Mechanism of action of _sprG1_-encoded type I toxins in Staphylococcus aureus: from membrane alterations to mesosome-like structures formation and bacterial lysis.
Fermon L, Burel A, Ostyn E, Dréano S, Bondon A, Chevance S, Pinel-Marie ML. Fermon L, et al. Front Microbiol. 2023 Oct 3;14:1275849. doi: 10.3389/fmicb.2023.1275849. eCollection 2023. Front Microbiol. 2023. PMID: 37854335 Free PMC article. - Comparative Analysis of Diverse Acetyltransferase-Type Toxin-Antitoxin Loci in Klebsiella pneumoniae.
Goh YX, Li P, Wang M, Djordjevic M, Tai C, Wang H, Deng Z, Chen Z, Ou HY. Goh YX, et al. Microbiol Spectr. 2022 Aug 31;10(4):e0032022. doi: 10.1128/spectrum.00320-22. Epub 2022 Jun 15. Microbiol Spectr. 2022. PMID: 35703555 Free PMC article. - Crystallization and X-ray analysis of all of the players in the autoregulation of the ataRT toxin-antitoxin system.
Jurėnas D, Van Melderen L, Garcia-Pino A. Jurėnas D, et al. Acta Crystallogr F Struct Biol Commun. 2018 Jul 1;74(Pt 7):391-401. doi: 10.1107/S2053230X18007914. Epub 2018 Jun 26. Acta Crystallogr F Struct Biol Commun. 2018. PMID: 29969102 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases