A highly divergent archaeo-eukaryotic primase from the Thermococcus nautilus plasmid, pTN2 - PubMed (original) (raw)
A highly divergent archaeo-eukaryotic primase from the Thermococcus nautilus plasmid, pTN2
Sukhvinder Gill et al. Nucleic Acids Res. 2014 Apr.
Abstract
We report the characterization of a DNA primase/polymerase protein (PolpTN2) encoded by the pTN2 plasmid from Thermococcus nautilus. Sequence analysis revealed that this protein corresponds to a fusion between an N-terminal domain homologous to the small catalytic subunit PriS of heterodimeric archaeal and eukaryotic primases (AEP) and a C-terminal domain related to their large regulatory subunit PriL. This unique domain configuration is not found in other virus- and plasmid-encoded primases in which PriS-like domains are typically fused to different types of helicases. PolpTN2 exhibited primase, polymerase and nucleotidyl transferase activities and specifically incorporates dNTPs, to the exclusion of rNTPs. PolpTN2 could efficiently prime DNA synthesis by the T. nautilus PolB DNA polymerase, suggesting that it is used in vivo as a primase for pTN2 plasmid replication. The N-terminal PriS-like domain of PolpTN2 exhibited all activities of the full-length enzyme but was much less efficient in priming cellular DNA polymerases. Surprisingly, the N-terminal domain possesses reverse transcriptase activity. We speculate that this activity could reflect an ancestral function of AEP proteins in the transition from the RNA to the DNA world.
Figures
Figure 1.
Sequence analysis of the N-terminal PriS-like domain of PolpTN2. (A) Domain organization of the PolpTN2 primase–polymerase. The four conserved motifs of the Prim–pol domain and the four cysteine residues predicted to be involved in the Fe–S cluster formation are indicated with green and red bars, respectively [Figure adapted from (17)]. (B) Alignment of PolpTN2 with with the sequences of S. solfataricus PriS (PDB ID: 1ZT2) and the PriS-like domain of ORF904 from S. islandicus plasmid pRN1 (PDB ID: 3M1M). The alignment is coloured according to sequence conservation (BLOSUM62 matrix). The secondary structure of PolpTN2 is indicated above the alignment and was predicted using Jpred and PsiPred. Green ellipses and grey arrows represent respectively α-helixes and β-strands characteristic to the canonical AEP fold (4), while red ellipses and blue arrows represent α-helixes and β-strands that are specific to PolpTN2. Negatively and positively charged residues that constitute the active site of PriS-like proteins are depicted with red and blue circles, respectively (note that the catalytic His residue present in the β5 strand in canonical AEP superfamily proteins is not conserved in PolpTN2 [crossed blue circle]). (C) Multiple sequence alignment of the predicted Fe–S cluster binding motif of PolpTN2 with those found in the large subunits of the archaeal and eukaryotic primases. The alignment is coloured according to sequence conservation (BLOSUM62 matrix). The four conserved Cys residues responsible for coordination of the Fe–S cluster are highlighted in red. The limits of the depicted motifs are indicated by the residue positions on each side of the alignment, with the total length of the protein given in parenthesis. Numbers between the motifs indicate the spacing between the corresponding motifs. The sequences are indicated with their GenBank identifiers followed by the corresponding organism name.
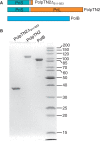
Figure 2.
PolpTN2Δ311–923, PolpTN2 and Thermococcus nautilus, PolB proteins. (A) Schematic representation of the polypeptides; see also Figure 1. (B) SDS-polyacrylamide gel analysis of purified PolpTN2Δ311–923 (0.5 µg), intact PolpTN2 (0.25 µg) and PolB (0.6 µg).
Figure 3.
Primase vs. primer-dependent DNA polymerase activity of intact PolpTN2, PolPTNΔ311–923 and Taq polymerase. (A) Intact PolpTN2 (40 ng/µl), (B) PolpTN2Δ311–923 (80 ng/µl) or (C) Taq DNA polymerase (0,05 u/µl) were incubated at 70°C in Taq buffer + 0,4 mM dNTP and 2 ng/µl M13mp18 DNA, which was or was not hybridized to the complementary oligonucleotide primer M13 forward (
Supplementary Table S1
). At t = 0, 5, 10 and 15 min, aliquots were withdrawn and quenched into 25 mM EDTA. The amount of ds DNA synthesized was then determined using Sybr® Green I fluorescence as described in Materials and Methods. Circles, continuous line: synthesis with hybridized primer; squares, dashed line: synthesis without primer. Points are the average of two determinations. The standard deviation is indicated by error bars.
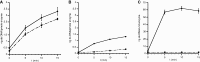
Figure 4.
Efficiency of intact PolpTN2 and PolpTN2Δ311–923 in priming DNA synthesis by T. nautilus PolB and Taq DNA polymerase. T. nautilus PolB (8 ng/µl) or Taq polymerase (0,05 u/µl) were incubated at 70°C in Taq buffer + 0,4 mM dNTP and 2 ng/µl M13mp18 DNA (without annealed primer) with increasing concentrations of either intact PolpTN2 (A) or PolpTN2 Δ311–923 (B). DNA synthesis by intact PolpTN2 or PolpTN2 Δ311–923 alone was included as a control. At t = 0 and 6 min, aliquots were withdrawn, quenched in 25 mM EDTA and assayed for ds DNA using Sybr® Green I fluorescence. Circles, continuous line: PolpTN2 or PolpTN2 Δ311–923 alone; squares, dashed line: PolpTN2 or PolpTN2 Δ311–923 plus Taq DNA polymerase; triangles, dotted line: PolpTN2 or PolpTN2 Δ311–923 plus T. nautilus PolB DNA polymerase. Points are the average of three determinations. The standard deviation is indicated by error bars.
Figure 5.
Strictly dNTP-dependent primase activity of PolpTN2 and PolpTN2Δ311–923. Primase reactions were performed in a total volume of 60 μl containing primase buffer, 5 μM of oligonucleotide (A45 [Spc C3]), 10 µM dNTPs, 10 µM NTPs, 0.6 nM [α-32P] dTTP and 1 µM of protein (PolpTN2 or PolpTN2Δ311–923). The reactions were incubated for 30 min at 70°C and subdivided into three 20-µl aliquots. The first aliquot was treated with 2 U of DNase, the second one with 0.3 M KOH and the third one received an equal volume of stop buffer [98% (v/v) formamide, 10 mM EDTA (pH 8), 1 mg/ml bromophenol blue]. The reaction products were then separated on a 16% denaturing polyacrylamide gel. Lane 1: aliquot treated with DNase; lane 2: aliquot treated with 0.3 M KOH; lane 3: aliquot left untreated. Markers are included on the left hand side of the gel (15-mer, 50-mer and 100-mer).
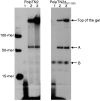
Figure 6.
Terminal transferase activity of PolpTN2 (A) and PolpTN2Δ311–923 (B). 5′ 32P-labelled substrates were incubated with PolpTN2 or PolpTN2Δ311–923 and non-labelled dNTPs in the presence of 10 mM of Mg2+ as described in Materials and Methods, followed by denaturing gel electrophoresis and autoradiography. A radiolabelled DNA ladder is included to the left and in the middle of the gel. The ladder also serves as a negative control as it consists of the templates used in the experiment. Templates consisted of 1: a single-stranded 52-mer (ss TT forward, see
Supplementary Table S1
) and 2: a double-stranded 52-mer (ss TT forward annealed to its complementary oligonucleotide ss TT reverse). Asterisks indicate the position of the 32P label.
Figure 7.
Reverse transcriptase activity of PolpTN2 and PolpTN2Δ311–923. A 30-mer RNA template (30 RT, see
Supplementary Table S1
) was annealed to a radiolabelled 20-mer primer (20 RT). Reactions were carried out in a volume of 20 µl of reverse transcriptase buffer. A control with commercial AMV enzyme was included. Reactions were quenched on ice and with addition of 20 µl of stop buffer containing formamide. 1 = PolpTN2, 2 = PolpTN2Δ311–923 and 3 = AMV commercial enzyme. The products longer than the template synthesized by PolpTN2Δ311–923 are probably due to the inherent terminal transferase activity of the enzyme.
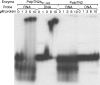
Figure 8.
Electrophoretic mobility shift assay. The RNA 30-mer 30 RT and its homologue 30 DNA were incubated with increasing concentrations of PolpTN2Δ311–923 and PolpTN2 as described in Materials and Methods. The enzyme, the type of probe and the concentration of each protein are indicated on top of the autoradiogram.
Similar articles
- Novel Families of Archaeo-Eukaryotic Primases Associated with Mobile Genetic Elements of Bacteria and Archaea.
Kazlauskas D, Sezonov G, Charpin N, Venclovas Č, Forterre P, Krupovic M. Kazlauskas D, et al. J Mol Biol. 2018 Mar 2;430(5):737-750. doi: 10.1016/j.jmb.2017.11.014. Epub 2017 Dec 11. J Mol Biol. 2018. PMID: 29198957 Free PMC article. - Synergistic template-free synthesis of dsDNA by Thermococcus nautili primase PolpTN2, DNA polymerase PolB, and pTN2 helicase.
Béguin P, Gill S, Charpin N, Forterre P. Béguin P, et al. Extremophiles. 2015 Jan;19(1):69-76. doi: 10.1007/s00792-014-0706-1. Epub 2014 Nov 25. Extremophiles. 2015. PMID: 25420601 - Characterization of DNA primase complex isolated from the archaeon, Thermococcus kodakaraensis.
Chemnitz Galal W, Pan M, Kelman Z, Hurwitz J. Chemnitz Galal W, et al. J Biol Chem. 2012 May 11;287(20):16209-19. doi: 10.1074/jbc.M111.338145. Epub 2012 Feb 17. J Biol Chem. 2012. PMID: 22351771 Free PMC article. - Initiating DNA replication: a matter of prime importance.
Bell SD. Bell SD. Biochem Soc Trans. 2019 Feb 28;47(1):351-356. doi: 10.1042/BST20180627. Epub 2019 Jan 15. Biochem Soc Trans. 2019. PMID: 30647143 Free PMC article. Review. - The Pol α-primase complex.
Pellegrini L. Pellegrini L. Subcell Biochem. 2012;62:157-69. doi: 10.1007/978-94-007-4572-8_9. Subcell Biochem. 2012. PMID: 22918585 Review.
Cited by
- A primase subunit essential for efficient primer synthesis by an archaeal eukaryotic-type primase.
Liu B, Ouyang S, Makarova KS, Xia Q, Zhu Y, Li Z, Guo L, Koonin EV, Liu ZJ, Huang L. Liu B, et al. Nat Commun. 2015 Jun 22;6:7300. doi: 10.1038/ncomms8300. Nat Commun. 2015. PMID: 26095544 Free PMC article. - MArVD2: a machine learning enhanced tool to discriminate between archaeal and bacterial viruses in viral datasets.
Vik D, Bolduc B, Roux S, Sun CL, Pratama AA, Krupovic M, Sullivan MB. Vik D, et al. ISME Commun. 2023 Aug 24;3(1):87. doi: 10.1038/s43705-023-00295-9. ISME Commun. 2023. PMID: 37620369 Free PMC article. - Primase-polymerases are a functionally diverse superfamily of replication and repair enzymes.
Guilliam TA, Keen BA, Brissett NC, Doherty AJ. Guilliam TA, et al. Nucleic Acids Res. 2015 Aug 18;43(14):6651-64. doi: 10.1093/nar/gkv625. Epub 2015 Jun 24. Nucleic Acids Res. 2015. PMID: 26109351 Free PMC article. Review. - Recent Mobility of Casposons, Self-Synthesizing Transposons at the Origin of the CRISPR-Cas Immunity.
Krupovic M, Shmakov S, Makarova KS, Forterre P, Koonin EV. Krupovic M, et al. Genome Biol Evol. 2016 Jan 13;8(2):375-86. doi: 10.1093/gbe/evw006. Genome Biol Evol. 2016. PMID: 26764427 Free PMC article. - Primer-Independent DNA Synthesis by a Family B DNA Polymerase from Self-Replicating Mobile Genetic Elements.
Redrejo-Rodríguez M, Ordóñez CD, Berjón-Otero M, Moreno-González J, Aparicio-Maldonado C, Forterre P, Salas M, Krupovic M. Redrejo-Rodríguez M, et al. Cell Rep. 2017 Nov 7;21(6):1574-1587. doi: 10.1016/j.celrep.2017.10.039. Cell Rep. 2017. PMID: 29117562 Free PMC article.
References
- Lipps G, Weinzierl AO, von Scheven G, Buchen C, Cramer P. Structure of a bifunctional DNA primase-polymerase. Nat. Struct. Mol. Biol. 2004;11:157–162. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials