Construction of synthetic nucleoli in human cells reveals how a major functional nuclear domain is formed and propagated through cell division - PubMed (original) (raw)
Construction of synthetic nucleoli in human cells reveals how a major functional nuclear domain is formed and propagated through cell division
Alice Grob et al. Genes Dev. 2014.
Abstract
Human cell nuclei are functionally organized into structurally stable yet dynamic bodies whose cell cycle inheritance is poorly understood. Here, we investigate the biogenesis and propagation of nucleoli, sites of ribosome biogenesis and key regulators of cellular growth. Nucleolar and cell cycles are intimately connected. Nucleoli disappear during mitosis, reforming around prominent uncharacterized chromosomal features, nucleolar organizer regions (NORs). By examining the effects of UBF depletion on both endogenous NORs and synthetic pseudo-NORs, we reveal its essential role in maintaining competency and establishing a bookmark on mitotic NORs. Furthermore, we demonstrate that neo-NORs, UBF-binding site arrays coupled with rDNA transcription units, direct the de novo biogenesis of functional compartmentalized neonucleoli irrespective of their site of chromosomal integration. For the first time, we establish the sequence requirements for nucleolar biogenesis and provide proof that this is a staged process where UBF-dependent mitotic bookmarking precedes function-dependent nucleolar assembly.
Keywords: UBF; mitotic bookmarking; neo-NOR; nucleolar organizer region (NOR); nucleolus; pseudo-NOR.
Figures
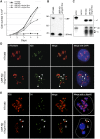
Figure 1.
UBF depletion induces NOR silencing. (A) Growth curves of HT1080 cells and UBF-KD cells cultured in the absence and presence of 2 ng/mL or 1 μg/mL Dox. (B) Western blot of proteins extracted from HT1080 cells and UBF-KD cells cultured with 2 ng/mL Dox. Molecular weight markers in kilodaltons are indicated at the left. Anti-UBF antibodies revealed a 6.5-fold depletion of UBF1 (97 kDa) and UBF2 (94 kDa), while anti-RPA43 antibodies reveal the conserved level of Pol I subunit RPA43 (43 kDa). (C) S1 nuclease protection assays performed on equal amounts of RNA extracted from HT1080 and UBF-KD cells indicate that the rDNA transcription level is conserved in UBF-KD cells cultured with 2 ng/mL Dox. S1 assays performed on equal amounts of RNA extracted from HT1080 and UBF-KD cells cultured in 2 ng/mL Dox reveal that UBF depletion does not affect the efficiency of cleavage at the A′ site in the 5′ ETS of pre-rRNA. Escherichia coli RNA served as a negative control. Protected fragments arising from cleaved (C) and uncleaved (UC) transcripts are indicated. (D) Combined 3D immuno-FISH performed on HT1080 and UBF-KD cells cultured with 2 ng/mL Dox reveals the dissociation of rDNA repeats (IGS probe) from the nucleolus (fibrillarin immunostaining) upon UBF depletion (arrowheads). (E) Combined 3D immuno-FISH performed on HT1080 and UBF-KD cells cultured with 2 ng/mL Dox reveals that foci of rDNA staining dissociated from nucleoli represent individual silent NORs (arrowheads). NORs are identified using a DJ BAC (green) that maps to the distal side of rDNA arrays, rDNA is identified using an IGS probe (red), and nucleoli are visualized by staining with anti-Nop52 antibodies (blue). Dashed lines indicate the edges of nuclei.
Figure 2.
Formation of secondary constriction on mitotic chromosomes is UBF-dependent. (A) Schematic representation of the novel secondary constriction formed by the insertion of a 1.4-Mb XEn array into the q-arm of chromosome 10 in pseudo-NOR clone 3D. (B) Western blot of proteins extracted from pseudo-NOR clone 3D cells transfected with control or UBF siRNAs revealed a 13-fold depletion of UBF1 and UBF2. (C) Metaphase chromosome spreads from pseudo-NOR clone 3D were hybridized with an XEn probe. Arrowheads indicate the site of pseudo-NOR insertion. Transfection of UBF siRNA induces the loss of pseudo-NOR secondary constriction. (D) Combined FISH and silver staining of metaphase spreads from pseudo-NOR clone 3D. UBF depletion results in the loss of silver staining at pseudo-NORs (arrowheads). Note the silver staining of an endogenous NOR (*) in the bottom right panel.
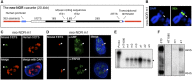
Figure 3.
Neo-NORs are undercondensed during mitosis and transcribed by Pol I into 45S pre-rRNA in interphase. (A) Schematic representation of the neo-NOR cassette. (B) Metaphase chromosome spreads from neo-NOR clone m1 were hybridized with an XEn probe. In clone m1, an array of neo-NOR cassettes is inserted into the p-arm of chromosome 4 (Supplemental Fig. S4). Arrowheads indicate novel secondary constrictions at the site of neo-NOR insertion. (C) RNA-FISH using mouse (green) and human (red) 5′ ETS probes identifies neo-NOR-derived transcripts (arrowheads) in clone m1. (D) Combined 3D immuno-RNA-FISH reveals that UBF and PAF49 colocalize with neo-NOR-derived transcripts in clone m1. (E) S1 nuclease protection assays on RNA extracted from HT1080 and neo-NOR cell lines (20 μg each) were performed at nucleotide resolution. These assays indicate that neo-NOR-derived transcripts initiate from the expected nucleotide +1 of neo-NOR 5′ ETS, resulting in the protection of 41 base pairs (bp) from the 60-bp probe. (F) Northern blotting of RNA extracted from 3T3 cells (1 μg), HT1080 cells (10 μg), and neo-NOR clones a1 and m1 (10 μg each) probed with mouse 5′ ETS sequences identifies neo-NOR-derived 45S pre-rRNAs and 34S pre-rRNA processing intermediates.
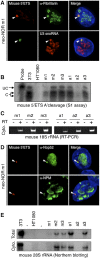
Figure 4.
Neonucleoli recruit DFC and GC nucleolar components and produce mature neo-NOR-derived rRNAs. (A) Combined 3D immuno-RNA-FISH reveals that early pre-rRNA processing factors fibrillarin and U3 snoRNA colocalize with neo-NOR-derived transcripts in clone m1. (B) S1 assays performed on RNA extracted from 3T3 (0.2 μg), HT1080 (20 μg), and neo-NOR (20 μg) cell lines indicate that neo-NOR-derived transcripts are accurately and efficiently cleaved at the A′ site of their 5′ ETS. Protected fragments arising from cleaved (C) and uncleaved (UC) transcripts are indicated. (C) RT–PCR reveals the presence of mouse 18S rRNAs in the cytoplasm of neo-NOR cell lines. Reactions in which reverse transcriptase (RT) was omitted served as negative controls. (D) Combined 3D immuno-RNA-FISH reveals that late pre-rRNA processing factors Nop52 and NPM colocalize with neo-NOR-derived transcripts in clone m1. (E) Northern blots of cellular (Total) and cytoplasmic (Cyto.) RNA extracted from 3T3 (0.5 μg), HT1080 (20 μg), and neo-NOR (20 μg) cell lines probed with a mouse-specific oligonucleotide reveal the presence of cytoplasmic mouse 28S rRNAs in neo-NOR lines.
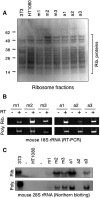
Figure 5.
Neo-NOR-derived rRNAs are assembled into polysome-associated ribosomes. (A) Coomassie-stained SDS protein gel of ribosome fractions purified from 3T3, HT1080, and neo-NOR cell lines. Ribosomal proteins are indicated. (B) RT–PCR detects neo-NOR-derived 18S rRNA in purified ribosome (Rib.) and polysome (Poly.) fractions. (C) Northern blots of RNA extracted from purified ribosomes (Rib.) and polysomes (Poly.) of 3T3 (0.5 μg), HT1080 (20 μg), and neo-NOR (20 μg) cell lines probed with a mouse-specific oligonucleotide indicate that neo-NOR-derived 28S rRNAs are incorporated into polysome-associated ribosomes within neo-NOR lines.
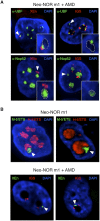
Figure 6.
Compartmentalized neonucleoli occupy distinct territories when integrated into endogenous nucleoli. (A) Combined 3D immuno-FISH performed on AMD-treated clone m1 reveals the presence of appropriately compartmentalized neonucleoli derived entirely from the neo-NOR. The top panels show UBF immunostained FC/DFC caps containing neo-NOR DNA (XEn probe; left panel) and lacking endogenous rDNA (IGS probe; right panel). The bottom panels show neonucleoli with a Nop52 immunostained GC associated with neo-NOR DNA (XEn probe; left panel) but lacking endogenous rDNA (IGS probe; right panel). Neonucleoli are indicated by arrowheads. In the enlarged insets, GCs, identified by low DAPI staining, are highlighted with a dashed line. (B, top panel) RNA-FISH performed on metacentric clone m1 identifies a distinct subnucleolar neo-NOR territory. Neo-NOR and endogenous transcripts are detected with mouse and human 5′ ETS probes, respectively. (Bottom panel) FISH performed on neo-NOR clone m1 indicates that neo-NORs and endogenous NORs are resolved into distinct nucleolar caps associated with the same GC upon AMD treatment. Neo-NORs and endogenous NORs were identified using XEn and IGS probes, respectively. GCs are identified by low DAPI staining. Images presented in the bottom panel correspond to single optical planes selected from deconvolved Z stacks.
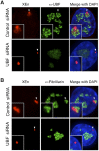
Figure 7.
Formation of neonucleoli is UBF-dependent. (A) Combined 3D immuno-FISH performed on neo-NOR clone m1 transfected with control or UBF siRNA. Note that the loss of UBF is associated with the compaction of neo-NOR sequences (XEn probe). (B) Combined 3D immuno-FISH performed on neo-NOR clone m1 transfected with control or UBF siRNA reveals that neo-NOR sequences (XEn probe) devoid of associated UBF fail to recruit fibrillarin. Further images from these experiments are shown in Supplemental Figure S11.
Comment in
- Driving nucleolar assembly.
McCann KL, Baserga SJ. McCann KL, et al. Genes Dev. 2014 Feb 1;28(3):211-3. doi: 10.1101/gad.237610.114. Genes Dev. 2014. PMID: 24493643 Free PMC article.
Similar articles
- Construction of synthetic nucleoli and what it tells us about propagation of sub-nuclear domains through cell division.
Grob A, McStay B. Grob A, et al. Cell Cycle. 2014;13(16):2501-8. doi: 10.4161/15384101.2014.949124. Cell Cycle. 2014. PMID: 25486191 Free PMC article. - Driving nucleolar assembly.
McCann KL, Baserga SJ. McCann KL, et al. Genes Dev. 2014 Feb 1;28(3):211-3. doi: 10.1101/gad.237610.114. Genes Dev. 2014. PMID: 24493643 Free PMC article. - The linker histone H1.2 is a novel component of the nucleolar organizer regions.
Chen J, Teo BHD, Cai Y, Wee SYK, Lu J. Chen J, et al. J Biol Chem. 2018 Feb 16;293(7):2358-2369. doi: 10.1074/jbc.M117.810184. Epub 2018 Jan 4. J Biol Chem. 2018. PMID: 29301938 Free PMC article. - Pseudo-NORs: a novel model for studying nucleoli.
Prieto JL, McStay B. Prieto JL, et al. Biochim Biophys Acta. 2008 Nov;1783(11):2116-23. doi: 10.1016/j.bbamcr.2008.07.004. Epub 2008 Jul 18. Biochim Biophys Acta. 2008. PMID: 18687368 Review. - Nucleolar organizer regions: genomic 'dark matter' requiring illumination.
McStay B. McStay B. Genes Dev. 2016 Jul 15;30(14):1598-610. doi: 10.1101/gad.283838.116. Genes Dev. 2016. PMID: 27474438 Free PMC article. Review.
Cited by
- Nucleolar URB1 ensures 3' ETS rRNA removal to prevent exosome surveillance.
Shan L, Xu G, Yao RW, Luan PF, Huang Y, Zhang PH, Pan YH, Zhang L, Gao X, Li Y, Cao SM, Gao SX, Yang ZH, Li S, Yang LZ, Wang Y, Wong CCL, Yu L, Li J, Yang L, Chen LL. Shan L, et al. Nature. 2023 Mar;615(7952):526-534. doi: 10.1038/s41586-023-05767-5. Epub 2023 Mar 8. Nature. 2023. PMID: 36890225 - NORs on human acrocentric chromosome p-arms are active by default and can associate with nucleoli independently of rDNA.
van Sluis M, van Vuuren C, Mangan H, McStay B. van Sluis M, et al. Proc Natl Acad Sci U S A. 2020 May 12;117(19):10368-10377. doi: 10.1073/pnas.2001812117. Epub 2020 Apr 24. Proc Natl Acad Sci U S A. 2020. PMID: 32332163 Free PMC article. - Nuclear organization mediates cancer-compromised genetic and epigenetic control.
Zaidi SK, Fritz AJ, Tracy KM, Gordon JA, Tye CE, Boyd J, Van Wijnen AJ, Nickerson JA, Imbalzano AN, Lian JB, Stein JL, Stein GS. Zaidi SK, et al. Adv Biol Regul. 2018 Aug;69:1-10. doi: 10.1016/j.jbior.2018.05.001. Epub 2018 May 9. Adv Biol Regul. 2018. PMID: 29759441 Free PMC article. Review. - Emergent microenvironments of nucleoli.
King MR, Ruff KM, Pappu RV. King MR, et al. Nucleus. 2024 Dec;15(1):2319957. doi: 10.1080/19491034.2024.2319957. Epub 2024 Mar 5. Nucleus. 2024. PMID: 38443761 Free PMC article. Review. - Imaging stress.
Brielle S, Gura R, Kaganovich D. Brielle S, et al. Cell Stress Chaperones. 2015 Nov;20(6):867-74. doi: 10.1007/s12192-015-0615-y. Epub 2015 Jul 4. Cell Stress Chaperones. 2015. PMID: 26139131 Free PMC article.
References
- Albert B, Colleran C, Leger-Silvestre I, Berger AB, Dez C, Normand C, Perez-Fernandez J, McStay B, Gadal O 2013. Structure–function analysis of Hmo1 unveils an ancestral organization of HMG-box factors involved in ribosomal DNA transcription from yeast to human. Nucleic Acids Res 41: 10135–10149 - PMC - PubMed
- Belin S, Hacot S, Daudignon L, Therizols G, Pourpe S, Mertani HC, Rosa-Calatrava M, Diaz JJ 2010. Purification of ribosomes from human cell lines. Curr Protoc Cell Biol 49: 3.40.1–3.40.11 - PubMed
- Boisvert FM, van Koningsbruggen S, Navascues J, Lamond AI 2007. The multifunctional nucleolus. Nat Rev Mol Cell Biol 8: 574–585 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources