Astrocytes play a key role in Drosophila mushroom body axon pruning - PubMed (original) (raw)
Astrocytes play a key role in Drosophila mushroom body axon pruning
Yaniv Hakim et al. PLoS One. 2014.
Abstract
Axon pruning is an evolutionarily conserved strategy used to remodel neuronal connections during development. The Drosophila mushroom body (MB) undergoes neuronal remodeling in a highly stereotypical and tightly regulated manner, however many open questions remain. Although it has been previously shown that glia instruct pruning by secreting a TGF-β ligand, myoglianin, which primes MB neurons for fragmentation and also later engulf the axonal debris once fragmentation has been completed, which glia subtypes participate in these processes as well as the molecular details are unknown. Here we show that, unexpectedly, astrocytes are the major glial subtype that is responsible for the clearance of MB axon debris following fragmentation, even though they represent only a minority of glia in the MB area during remodeling. Furthermore, we show that astrocytes both promote fragmentation of MB axons as well as clear axonal debris and that this process is mediated by ecdysone signaling in the astrocytes themselves. In addition, we found that blocking the expression of the cell engulfment receptor Draper in astrocytes only affects axonal debris clearance. Thereby we uncoupled the function of astrocytes in promoting axon fragmentation to that of clearing axonal debris after fragmentation has been completed. Our study finds a novel role for astrocytes in the MB and suggests two separate pathways in which they affect developmental axon pruning.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
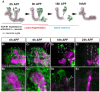
Figure 1. Astrocytes surround the MB during developmental axon pruning.
(A) Scheme of developmental pruning of MB γ neurons. During larval development and up to puparium formation (0 h APF), γ neurons extend a single process that sends out dendrites near the cell body and continues as an axon peduncle that bifurcates to form a dorsal and a medial branch. At the onset of puparium, a glial derived TGF-β signal induces the expression of the ecdysone receptor-B1 (EcR-B1) within γ neurons. Subsequently, an ecdysone pulse activates EcR-B1, resulting in a largely unknown transcriptional cascade. At 6 h APF both axonal branches, as well as the dendrites begin to undergo fragmentation, while the pedunclar axon remains intact and the neurons retain their cell body. The fragmented axons are then cleared by glia that surround these axons. Subsequently, γ neurons regrow axons that project into a new, adult specific and medially projecting lobe. (B–E) Confocal Z-projections of brains expressing alrm-GAL4 driving mCD8::GFP at 0 h APF (B), 6 h APF (C), 18 h APF (D) or 24 h APF (E). (B) At the onset of metamorphosis (0 h APF) astrocytic membranes are evenly dispersed in the region of the MB lobes (higher magnification in A2). (C) By 6 h APF, at the onset of pruning, the astrocytes have changed their morphology and have begun to infiltrate the degenerating lobes. (D) At 18 h astrocyte membranes surround axon fragments (arrowhead in D2). By 18 h to (E) 24 h APF there is a significant decrease in alrm-GAL4 positive membranes. Newly extended axon branches of α/β neurons, are also stained with anti-FasII antibody at 24 h APF (E). Magenta represents anti-FasII staining. Green is alrm-GAL4 driven mCD8::GFP. The scale bars are 20 μm. Genotypes: (B–E) y,w;alrm-GAL4;alrm-GAL4,UAS-mCD8::GFP/+.
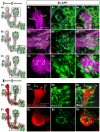
Figure 2. Astrocytes infiltrate and engulf degenerating MB γ neurons at 6h APF.
(A–C) Confocal Z-projections of brains expressing alrm-GAL4 driven mCD8::GFP focusing on the MB medial (A) or dorsal (B) lobes or the peduncle (C) at 6 h APF as depicted by the cartoons on the left (A1–C1). Fas II staining shows clear spherical holes devoid of FasII expression in the MB γ dorsal (A2) and medial (B2) lobes and a slight decrease in FasII staining in the peduncle (C3). These holes are occupied by astrocyte extensions (A3–C3) infiltrating into the MB axon bundle (arrows in A4–C4). Magenta represents anti-FasII staining. Green is alrm-GAL4 driven mCD8::GFP. (D-E) Confocal single slices of the MB dorsal lobe tip (D) and the peduncle (E). While γ neurons were labeled by the Q-system (ET40-QF driving QUAS-mtdT-3xHA expression), astrocytes were labeled by alrm-GAL4 driven mCD8::GFP expression. At 6 h APF, γ axons are largely fragmented at the dorsal tip lobe (D2) but only few fragments are detected at the peduncle (E2). While astrocytes infiltrate both the dorsal lobe (D4) and the peduncle (E4), clear engulfment events can only be seen around the dorsal tip (D4). Red is QF-ET40 driven QUAS-mtdt-3xHA. Green is alrm-GAL4 driven mCD8::GFP. The scale bars are 20 μm. Genotypes: (A–C) w;alrm-GAL4;alrm-GAL4,UAS-mCD8::GFP/+ (D–E) y,w;QF-ET40,QUAS-mtdT-3xHA/+;alrm-GAL4,UAS-mCD8::GFP/+.
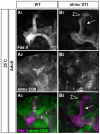
Figure 3. The number of astrocytes at the MB remains constant throughout remodeling.
(A–E) Confocal Z-projections of alrm-GAL4 driven RedStinger expression (red, A–E) labeling astrocytes nuclei at 0 h APF (A), 6 h APF (B, E), 18 h APF (C) or 24 h APF (D). (A–D) Labeling astrocyte cell bodies shows that the number of the astrocytes does not significantly change during remodeling. (E) High magnification view of dorsal tip reveals that astrocytes (red) label only part of the repo+ nuclei in the vicinity of the MB lobe tip. (F) To characterize this quantitatively, we counted the number of astrocytes nuclei within 5 μM of the dorsal tip and found that only 1–2 astrocytes are located near the dorsal tip throughout remodeling. Magenta represents anti-FasII and anti-Repo staining. Red is alrm-GAL4 driven RedStinger. (G) Confocal Z-projections (G) or single slices (individual cells) of brains expressing alrm-Gal4 driven CD8-GFP in just a few cells (see materials and methods). While the 6 glial cells proximal to the dorsal tip are not labeled with mCD8::GFP (close-up of dorsal tip, region 5), four cells that are located further away from the lobe are (see close-ups of cell 1–4). Grey represents FasII and Repo staining (G1 and upper panels in close-ups), DAPI staining in the lower panels in close ups or alrm-Gal4 driven CD8-GFP (G2 and middle panels in close-ups). Magenta represents FasII and Repo staining and green represents alrm-Gal4 driven CD8-GFP in H3 and lower panels in close-ups. The scales bars are 20 μm. Genotypes: (A–E): w;UAS-RedStinger/+;alrm-GAL4/+. (G) y,w,hsFlp,FRT19A/y,w,hsFlp,19A,tubP-GAL80;alrm-GAL4;alrm-GAL4,UAS-mCD8::GFP.
Figure 4. Astrocytes are necessary for efficient axon pruning.
(A–B) Confocal Z-projections of adult brains expressing CD8-GFP (A) or additionally UAS-DTI (diphtheria toxin; B) driven by alrm-Gal4. Driving the expression of DTI in astrocytes resulted in their partial ablation (grey, A2, B2) and results in fragmentation defects (arrow in B1) and uncleared debris (arrowhead in B1) in escapers (13 out of 118 expected flies). Grey in A1–B1 and magenta in A3–B3 represents anti-FasII. Grey in A2–B2 and green in A3–B3 are alrm-GAL4 driven mCD8::GFP. The scale bar is 20 μm. Genotypes: (A) y,w;Sp/CyO;alrm-GAL4,UAS-mCD8::GFP/+; (B) y,w;UAS-DTI/+;alrm GAL4, UAS-mCD8::GFP/+.
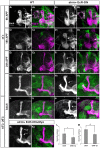
Figure 5. Astrocyte's Ecdysone Receptor controls multiply aspects of MB γ neuron remodeling.
Confocal Z-projections of brains expressing CD8-GFP driven by alrm-Gal4 (A, C, E, G, I), or those additionally expressing EcR-DN (B, D, F, H, J) or myo in addition to EcR-DN (K) in astrocytes at 6 h APF (A, B), 18 h APF (C, D), 24 h APF (E, F) and Adult (G–K) reared at either 25°C (A–H) or reared at 18°C until late larval stage and then transferred to 29°C until eclosion (see Fig. S2B, I–K). (A,B) At 6 h APF astrocytes expressing EcR-DN do not infiltrate the MB γ lobes (see insets). Overexpression of EcR in astrocytes results in a delay in axon pruning at 18 h APF (D compare to C) and at 24 h APF (F compare to E) that persisted into adult (H compare to G). Elevated expression in astrocytes (I–J; see S2A for details) resulted in more persistent unpruned axons. Overexpression of Myo in addition to EcR-DN (K) partially rescued both the fragmentation (L) and the debris clearance (M) defects. Quantification was performed by ranking analyses that are detailed in the materials and methods section. * P<0.05; ** P<0.005. Bars represent SEM. Grey in A1–K1 and magenta in A2–K2 represents anti-FasII staining. Green in A2–K2 represents alrm-GAL4 driven mCD8::GFP. Scale bar is 20 μM. Genotypes: (A–I): y,w; alrm-GAL4/+;alrm-GAL4,mCD8::GFP/+ (B–J): y,w; alrm-GAL4/UAS-EcR-DN;alrm-GAL4,mCD8::GFP/+. (K): y,w; alrm-GAL4/UAS-EcR-DN;alrm-GAL4,mCD8::GFP/UAS-Myo.
Figure 6. Knocking down the ecdysone regulated drpr or inhibiting endocytosis in astrocytes results in uncleared axonal debris.
Confocal Z-projections of adult (A–D) or 6APF brains (E, F) expressing CD8-GFP (A, E), additionally expressing Drpr-RNAi (B, C), Shits1 (D), or EcR-DN (F) in astrocytes using the alrm-Gal4 driver. High and prolonged expression of Drpr-RNAi (C) was achieved by the Tub>GAL4 flipout system (see Fig. S3A for details) (A2–D2) Represent a higher-magnification of the dorsal tip. No ectopic γ axon branches were detected in adults when astrocytic engulfment ability was impaired with expression of Drpr RNAi (B1, C1) or shibirets (D1). Uncleared debris (arrowhead) was detected in 100% of flies of the tested transgenes (Drpr RNAi, 12 flies, B2; forced expressed Drpr RNAi, 12 flies, C2 and shibirets, 17 flies, D2). (E, F) Drpr staining in WT (E) and flies expressing EcR-DN in astrocytes (F) at 6APF. Marked box represents regions that were used fort the quantification in G. (G) EcR-DN expressing flies show significantly lower staining for Drpr (0.16±0.01, n = 14) than WT flies (12.88±1.37, n = 10, p<0.001) at 6APF (quantification was done on confocal single slices). Grey represent anti-FasII staining in A–D and in E1–F1 and Drpr staining in E2–F2. Green in E and F represents alrm-GAL4 driven mCD8::GFP. Red in E3–4 and F3–4 is Drpr staining. The scale bars are 20 μm. Genotypes: (A) y,w;alrm-GAL4/+;alrm-GAL4,mCD8::GFP/+; (B) y,w;alrm-GAL4/+;alrm-GAL4,mCD8::GFP,Dcr2/UAS-Drpr-RNAi; (C) y,w;P{GAL4-αTub84B(FRT.CD2).P}/UAS-FLP;alrm-GAL4,mCD8::GFP,Dcr2/UAS-Drpr-RNA (D) y,w;alrm-GAL4/+;alrm-GAL4,mCD8::GFP/UAS-Shitts. (E) y,w; alrm-GAL4/+;alrm-GAL4,mCD8::GFP/+ (F) y,w; alrm-GAL4/UAS-EcR-DN;alrm-GAL4,mCD8::GFP/+.
Similar articles
- Astrocytes engage unique molecular programs to engulf pruned neuronal debris from distinct subsets of neurons.
Tasdemir-Yilmaz OE, Freeman MR. Tasdemir-Yilmaz OE, et al. Genes Dev. 2014 Jan 1;28(1):20-33. doi: 10.1101/gad.229518.113. Epub 2013 Dec 20. Genes Dev. 2014. PMID: 24361692 Free PMC article. - Genomic analysis of Drosophila neuronal remodeling: a role for the RNA-binding protein Boule as a negative regulator of axon pruning.
Hoopfer ED, Penton A, Watts RJ, Luo L. Hoopfer ED, et al. J Neurosci. 2008 Jun 11;28(24):6092-103. doi: 10.1523/JNEUROSCI.0677-08.2008. J Neurosci. 2008. PMID: 18550751 Free PMC article. - Engulfing action of glial cells is required for programmed axon pruning during Drosophila metamorphosis.
Awasaki T, Ito K. Awasaki T, et al. Curr Biol. 2004 Apr 20;14(8):668-77. doi: 10.1016/j.cub.2004.04.001. Curr Biol. 2004. PMID: 15084281 - Axon pruning: an active role for glial cells.
Broadie K. Broadie K. Curr Biol. 2004 Apr 20;14(8):R302-4. doi: 10.1016/j.cub.2004.03.049. Curr Biol. 2004. PMID: 15084298 Review. No abstract available. - Spatiotemporal regulation of developmental neurite pruning: Molecular and cellular insights from Drosophila models.
Furusawa K, Emoto K. Furusawa K, et al. Neurosci Res. 2021 Jun;167:54-63. doi: 10.1016/j.neures.2020.11.010. Epub 2020 Dec 10. Neurosci Res. 2021. PMID: 33309868 Review.
Cited by
- Analysis of Glial Distribution in Drosophila Adult Brains.
Ou J, Gao Z, Song L, Ho MS. Ou J, et al. Neurosci Bull. 2016 Apr;32(2):162-70. doi: 10.1007/s12264-016-0014-0. Epub 2016 Jan 25. Neurosci Bull. 2016. PMID: 26810782 Free PMC article. - Nitric Oxide as a Switching Mechanism between Axon Degeneration and Regrowth during Developmental Remodeling.
Rabinovich D, Yaniv SP, Alyagor I, Schuldiner O. Rabinovich D, et al. Cell. 2016 Jan 14;164(1-2):170-182. doi: 10.1016/j.cell.2015.11.047. Cell. 2016. PMID: 26771490 Free PMC article. - Astrocytes engage unique molecular programs to engulf pruned neuronal debris from distinct subsets of neurons.
Tasdemir-Yilmaz OE, Freeman MR. Tasdemir-Yilmaz OE, et al. Genes Dev. 2014 Jan 1;28(1):20-33. doi: 10.1101/gad.229518.113. Epub 2013 Dec 20. Genes Dev. 2014. PMID: 24361692 Free PMC article. - Mechanisms of developmental neurite pruning.
Schuldiner O, Yaron A. Schuldiner O, et al. Cell Mol Life Sci. 2015 Jan;72(1):101-19. doi: 10.1007/s00018-014-1729-6. Epub 2014 Sep 12. Cell Mol Life Sci. 2015. PMID: 25213356 Free PMC article. Review. - Glial cells in neuronal development: recent advances and insights from Drosophila melanogaster.
Ou J, He Y, Xiao X, Yu TM, Chen C, Gao Z, Ho MS. Ou J, et al. Neurosci Bull. 2014 Aug;30(4):584-94. doi: 10.1007/s12264-014-1448-2. Neurosci Bull. 2014. PMID: 25015062 Free PMC article. Review.
References
- Luo L, O'Leary DD (2005) Axon retraction and degeneration in development and disease. Annu Rev Neurosci 28: 127–156. - PubMed
- Saxena S, Caroni P (2007) Mechanisms of axon degeneration: from development to disease. Prog Neurobiol 83: 174–191. - PubMed
- Lee T, Lee A, Luo L (1999) Development of the Drosophila mushroom bodies: sequential generation of three distinct types of neurons from a neuroblast. Development 126: 4065–4076. - PubMed
- Zheng X, Wang J, Haerry TE, Wu AY, Martin J, et al. (2003) TGF-beta signaling activates steroid hormone receptor expression during neuronal remodeling in the Drosophila brain. Cell 112: 303–315. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
This work was funded by the Israel Science Foundation grants (ISF) 1864/08 and 683/11 (Bio-med Legacy program) and the Yeda-Sela Foundation. All the images were obtained using a Zeiss LSM710 confocal microscope that was purchased with the help of the Adelis Foundation. O.S. is incumbent of the Aser Rothstein Career Development Chair. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous