Design and characterization of a 52K SNP chip for goats - PubMed (original) (raw)
. 2014 Jan 22;9(1):e86227.
doi: 10.1371/journal.pone.0086227. eCollection 2014.
Philippe Bardou 2, Olivier Bouchez 3, Cédric Cabau 2, Richard Crooijmans 4, Yang Dong 5, Cécile Donnadieu-Tonon 3, André Eggen 6, Henri C M Heuven 7, Saadiah Jamli 8, Abdullah Johari Jiken 8, Christophe Klopp 9, Cynthia T Lawley 6, John McEwan 10, Patrice Martin 11, Carole R Moreno 12, Philippe Mulsant 1, Ibouniyamine Nabihoudine 2, Eric Pailhoux 13, Isabelle Palhière 12, Rachel Rupp 12, Julien Sarry 1, Brian L Sayre 14, Aurélie Tircazes 12, Jun Wang 15, Wen Wang 16, Wenguang Zhang 17; International Goat Genome Consortium
Collaborators, Affiliations
- PMID: 24465974
- PMCID: PMC3899236
- DOI: 10.1371/journal.pone.0086227
Design and characterization of a 52K SNP chip for goats
Gwenola Tosser-Klopp et al. PLoS One. 2014.
Erratum in
- Correction: Design and Characterization of a 52K SNP Chip for Goats.
Tosser-Klopp G, Bardou P, Bouchez O, Cabau C, Crooijmans R, Dong Y, Donnadieu-Tonon C, Eggen A, Heuven HC, Jamli S, Jiken AJ, Klopp C, Lawley CT, McEwan J, Martin P, Moreno CR, Mulsant P, Nabihoudine I, Pailhoux E, Palhière I, Rupp R, Sarry J, Sayre BL, Tircazes A, Wang J, Wang W, Zhang W; International Goat Genome Consortium. Tosser-Klopp G, et al. PLoS One. 2016 Mar 24;11(3):e0152632. doi: 10.1371/journal.pone.0152632. eCollection 2016. PLoS One. 2016. PMID: 27011020 Free PMC article. No abstract available.
Abstract
The success of Genome Wide Association Studies in the discovery of sequence variation linked to complex traits in humans has increased interest in high throughput SNP genotyping assays in livestock species. Primary goals are QTL detection and genomic selection. The purpose here was design of a 50-60,000 SNP chip for goats. The success of a moderate density SNP assay depends on reliable bioinformatic SNP detection procedures, the technological success rate of the SNP design, even spacing of SNPs on the genome and selection of Minor Allele Frequencies (MAF) suitable to use in diverse breeds. Through the federation of three SNP discovery projects consolidated as the International Goat Genome Consortium, we have identified approximately twelve million high quality SNP variants in the goat genome stored in a database together with their biological and technical characteristics. These SNPs were identified within and between six breeds (meat, milk and mixed): Alpine, Boer, Creole, Katjang, Saanen and Savanna, comprising a total of 97 animals. Whole genome and Reduced Representation Library sequences were aligned on >10 kb scaffolds of the de novo goat genome assembly. The 60,000 selected SNPs, evenly spaced on the goat genome, were submitted for oligo manufacturing (Illumina, Inc) and published in dbSNP along with flanking sequences and map position on goat assemblies (i.e. scaffolds and pseudo-chromosomes), sheep genome V2 and cattle UMD3.1 assembly. Ten breeds were then used to validate the SNP content and 52,295 loci could be successfully genotyped and used to generate a final cluster file. The combined strategy of using mainly whole genome Next Generation Sequencing and mapping on a contig genome assembly, complemented with Illumina design tools proved to be efficient in producing this GoatSNP50 chip. Advances in use of molecular markers are expected to accelerate goat genomic studies in coming years.
Conflict of interest statement
Competing Interests: André Eggen and Cynthia T. Lawley are employees of Illumina, Inc. This work led to the development of a goat 50K SNP chip, sold by Illumina to any customer. This does not alter the authors' adherence to all the PLOS ONE policies on sharing data and materials.
Figures
Figure 1. Goat Genome scaffolds assembly.
The goat genome scaffolds were sorted by decreasing size (x-axis) and the cumulative proportion of the assembled genome was plotted on the y-axis for all the scaffolds. The vertical line shows that >10kb scaffolds represent 97.2% of the assembled goat genome.
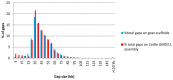
Figure 2. SNP spacing on the goat scaffolds.
Spacing between the selected SNPs was calculated and the percentage of gaps (total number of gaps is 59,030 on goat scaffolds and 62,693 on UMD3.1 cattle assembly) is shown (y-axis) in each 5kb class ranging from 5 to 150kb (x-axis).
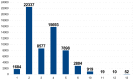
Figure 3. SNPs by category in final design.
The number of selected SNPs is indicated for each of the following categories. 1: SNP detected in an EST. 2: two alleles detected in the five considered breeds. 3: two alleles detected in Alpine and Saanen and Creole and (Boer or Savanna). 4: two alleles detected in two of the three milk and mixed breeds (Alpine, Saanen, Creole) and in Boer and Savanna. 5: two alleles detected in Alpine and Saanen and Creole. 6: two alleles detected in three out of the five breeds. 10: two alleles detected in each of the two milk breeds (Saanen and Alpine). 11: two alleles detected in one milk breed (Saanen or Alpine) and one meat breed (Creole or Boer or Katjang/Savanna). 12: two alleles detected in at least two meat breeds (Creole and Boer or Katjang/Savanna). 13: two alleles detected in one milk breed (Saanen or Alpine).
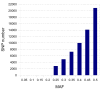
Figure 4. Distribution of estimated MAFs of the selected SNPs.
The MAF for all the 60,000 selected SNPs was estimated based on the read counts for the two alleles.
Similar articles
- The development and characterization of a 60K SNP chip for chicken.
Groenen MA, Megens HJ, Zare Y, Warren WC, Hillier LW, Crooijmans RP, Vereijken A, Okimoto R, Muir WM, Cheng HH. Groenen MA, et al. BMC Genomics. 2011 May 31;12(1):274. doi: 10.1186/1471-2164-12-274. BMC Genomics. 2011. PMID: 21627800 Free PMC article. - Design and validation of high-density SNP array of goats and population stratification of Indian goat breeds.
Vijh RK, Sharma U, Kapoor P, Raheja M, Arora R, Ahlawat S, Dureja V. Vijh RK, et al. Gene. 2023 Nov 15;885:147691. doi: 10.1016/j.gene.2023.147691. Epub 2023 Aug 5. Gene. 2023. PMID: 37544337 - Genome-wide association mapping for type and mammary health traits in French dairy goats identifies a pleiotropic region on chromosome 19 in the Saanen breed.
Martin P, Palhière I, Maroteau C, Clément V, David I, Klopp GT, Rupp R. Martin P, et al. J Dairy Sci. 2018 Jun;101(6):5214-5226. doi: 10.3168/jds.2017-13625. Epub 2018 Mar 21. J Dairy Sci. 2018. PMID: 29573797 - [Progress in goat genome studies].
Wang FH, Zhang L, Li XK, Fan YX, Qiao X, Gong G, Yan XC, Zhang LT, Wang ZY, Wang RJ, Liu ZH, Wang ZX, He L, Zhang Y, Li JQ, Zhao YH, Su R. Wang FH, et al. Yi Chuan. 2019 Oct 20;41(10):928-938. doi: 10.16288/j.yczz.19-147. Yi Chuan. 2019. PMID: 31624055 Review. Chinese. - Trajectory of livestock genomics in South Asia: A comprehensive review.
Panigrahi M, Kumar H, Saravanan KA, Rajawat D, Sonejita Nayak S, Ghildiyal K, Kaisa K, Parida S, Bhushan B, Dutt T. Panigrahi M, et al. Gene. 2022 Nov 15;843:146808. doi: 10.1016/j.gene.2022.146808. Epub 2022 Aug 13. Gene. 2022. PMID: 35973570 Review.
Cited by
- A genome-wide analysis of copy number variation in Murciano-Granadina goats.
Guan D, Martínez A, Castelló A, Landi V, Luigi-Sierra MG, Fernández-Álvarez J, Cabrera B, Delgado JV, Such X, Jordana J, Amills M. Guan D, et al. Genet Sel Evol. 2020 Aug 8;52(1):44. doi: 10.1186/s12711-020-00564-4. Genet Sel Evol. 2020. PMID: 32770942 Free PMC article. - Candidate Genes and Their Expressions Involved in the Regulation of Milk and Meat Production and Quality in Goats (Capra hircus).
Salgado Pardo JI, Delgado Bermejo JV, González Ariza A, León Jurado JM, Marín Navas C, Iglesias Pastrana C, Martínez Martínez MDA, Navas González FJ. Salgado Pardo JI, et al. Animals (Basel). 2022 Apr 11;12(8):988. doi: 10.3390/ani12080988. Animals (Basel). 2022. PMID: 35454235 Free PMC article. Review. - Genome-Wide Association Studies Identify Candidate Genes for Coat Color and Mohair Traits in the Iranian Markhoz Goat.
Nazari-Ghadikolaei A, Mehrabani-Yeganeh H, Miarei-Aashtiani SR, Staiger EA, Rashidi A, Huson HJ. Nazari-Ghadikolaei A, et al. Front Genet. 2018 Apr 4;9:105. doi: 10.3389/fgene.2018.00105. eCollection 2018. Front Genet. 2018. PMID: 29670642 Free PMC article. - Genetic diversity and signatures of selection in various goat breeds revealed by genome-wide SNP markers.
Brito LF, Kijas JW, Ventura RV, Sargolzaei M, Porto-Neto LR, Cánovas A, Feng Z, Jafarikia M, Schenkel FS. Brito LF, et al. BMC Genomics. 2017 Mar 14;18(1):229. doi: 10.1186/s12864-017-3610-0. BMC Genomics. 2017. PMID: 28288562 Free PMC article. - Wattles in goats are associated with the FMN1/GREM1 region on chromosome 10.
Reber I, Keller I, Becker D, Flury C, Welle M, Drögemüller C. Reber I, et al. Anim Genet. 2015 Jun;46(3):316-20. doi: 10.1111/age.12279. Epub 2015 Mar 3. Anim Genet. 2015. PMID: 25736034 Free PMC article.
References
- Taberlet P, Coissac E, Pansu J, Pompanon F (2011) Conservation genetics of cattle, sheep, and goats. C R Biol 334: 247–254. - PubMed
- Schibler L, Vaiman D, Oustry A, Guinec N, Dangy-Caye AL, et al. (1998) Construction and extensive characterization of a goat bacterial artificial chromosome library with threefold genome coverage. Mamm Genome 9: 119–124. - PubMed
- Schibler L, Vaiman D, Oustry A, Giraud-Delville C, Cribiu EP (1998) Comparative gene mapping: a fine-scale survey of chromosome rearrangements between ruminants and humans. Genome Res 8: 901–915. - PubMed
- Pailhoux E, Vigier B, Chaffaux S, Servel N, Taourit S, et al. (2001) A 11.7-kb deletion triggers intersexuality and polledness in goats. Nat Genet 29: 453–458. - PubMed
- Vaiman D, Koutita O, Oustry A, Elsen JM, Manfredi E, et al. (1996) Genetic mapping of the autosomal region involved in XX sex-reversal and horn development in goats. Mamm Genome 7: 133–137. - PubMed
Publication types
MeSH terms
Grants and funding
Grants were funded by ANR (http://www.agence-nationale-recherche.fr/), ANR-09-GENM-009-03 GENIDOV, CHEST-454, funded by APIS-GENE; CAPRISNIP programme: UNCEIA, CAPGENES and APIS-GENE French Breeding organizations; and EC’s Seventh Framework Programme, 3SR Integrated Project(Sustainable Solutions for Small Ruminants) http://www.3srbreeding.eu). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
LinkOut - more resources
Full Text Sources
Other Literature Sources