Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays - PubMed (original) (raw)
Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays
Martin J Aryee et al. Bioinformatics. 2014.
Abstract
Motivation: The recently released Infinium HumanMethylation450 array (the '450k' array) provides a high-throughput assay to quantify DNA methylation (DNAm) at ∼450 000 loci across a range of genomic features. Although less comprehensive than high-throughput sequencing-based techniques, this product is more cost-effective and promises to be the most widely used DNAm high-throughput measurement technology over the next several years.
Results: Here we describe a suite of computational tools that incorporate state-of-the-art statistical techniques for the analysis of DNAm data. The software is structured to easily adapt to future versions of the technology. We include methods for preprocessing, quality assessment and detection of differentially methylated regions from the kilobase to the megabase scale. We show how our software provides a powerful and flexible development platform for future methods. We also illustrate how our methods empower the technology to make discoveries previously thought to be possible only with sequencing-based methods.
Availability and implementation: http://bioconductor.org/packages/release/bioc/html/minfi.html.
Contact: khansen@jhsph.edu; rafa@jimmy.harvard.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
Figures
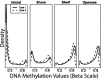
Fig. 1.
Beta density estimates for a typical sample showing type I (solid) and type II (dashed) loci located in CGIs, CGI shores, CGI shelves and open sea regions
Fig. 2.
Illustration of locus-collapsing procedure for block finding. Loci in CpG islands, shores, shelves and open sea regions are represented by green, orange, purple and pink, respectively. (A) The boxes represent locus groups, each of which is collapsed to a single mean methylation value. We group loci within the same CGI, the same CGI shore or the same CGI shelf, as well as adjacent open sea probes that are within 500 bp of each other. (B) The first row of points shows the midpoints of collapsed open sea clusters. These are grouped into long-range clusters and used for block finding. The second row of points shows all collapsed clusters across all region types with color representing region type
Fig. 3.
Accuracy and precision assessment of preprocessing algorithms. (A) For each locus, we compute the average and standard deviation across liver technical samples. The resulting loess curve fitted to the standard deviation versus average scatterplot for each method is shown. (B) Using the same samples, we compute the average difference between liver and placenta (effect size) for each locus. We then plot the resulting effect sizes for each preprocessing method against effect sizes from the default Illumina procedure
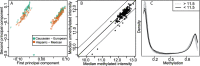
Fig. 4.
Quality assessment plots based on the blood sample dataset. (A) A multidimensional scaling plot. Color represents reported ethnicity. (B) Scatterplot of median Unmeth signal versus median Meth signal value for each sample. Points outside the dashed lines represent cases were the differences are >0.5. (C) Beta density plots for all samples with black curves representing samples where the average of the median Unmeth and Meth is <11.5
Fig. 5.
DMRs associate more strongly with gene expression than methylation differences at single CpGs, as observed in a dataset of normal lung and colon samples. (A) An example of a tissue-DMR, identified by bumphunter. The 15 CpGs in the region show concordant methylation differences. (B) An example of a significant tissue-DMP, identified by a locus-level limma model. Note that the CpG probes adjacent to the DMP do not show a methylation difference. (C) Between-tissue differential expression is greater for genes with a DMR located within 2 kb of the transcriptional start site (left) than for genes with a DMP located within 2 kb of the transcriptional start site (right). (D) A greater fraction of DMRs is located close to DEG promoters than are DMPs
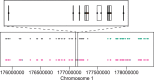
Fig. 6.
Large regions of hypomethylation in colon cancer are reliably identified by minfi. We used the block finding method on 450k data for colon cancer and matched normal samples from the TCGA project. The top (A) shows smoothed estimates of average methylation at the collapsed locus level in the region plotted as Figure 2a in Hansen et al. (2011). Loss of methylation in tumor is clearly observed in this region. The second panel shows the methylation difference between cancer and normal. Dots indicate the probe clusters used in the block finder algorithm, which ignores clusters corresponding to CpG islands, shores or shelves. The smoothed methylation difference used for segmentation is also plotted. The gap in this smooth curve results from large genomic distances between probe clusters over which no smoothing is performed. The bottom panel shows the minfi segmentation of the cluster-level measurements, with blue indicating blocks of significant hypomethylation. The bottom track are the blocks of methylation difference defined from whole-genome bisulfite sequencing in Hansen et al. (2011) (B) Hypomethylation regions identified by minfi consistently overlap hypomethylation blocks identified in Hansen et al. (2011)
Similar articles
- Preprocessing, normalization and integration of the Illumina HumanMethylationEPIC array with minfi.
Fortin JP, Triche TJ Jr, Hansen KD. Fortin JP, et al. Bioinformatics. 2017 Feb 15;33(4):558-560. doi: 10.1093/bioinformatics/btw691. Bioinformatics. 2017. PMID: 28035024 Free PMC article. - ChAMP: updated methylation analysis pipeline for Illumina BeadChips.
Tian Y, Morris TJ, Webster AP, Yang Z, Beck S, Feber A, Teschendorff AE. Tian Y, et al. Bioinformatics. 2017 Dec 15;33(24):3982-3984. doi: 10.1093/bioinformatics/btx513. Bioinformatics. 2017. PMID: 28961746 Free PMC article. - missMethyl: an R package for analyzing data from Illumina's HumanMethylation450 platform.
Phipson B, Maksimovic J, Oshlack A. Phipson B, et al. Bioinformatics. 2016 Jan 15;32(2):286-8. doi: 10.1093/bioinformatics/btv560. Epub 2015 Sep 30. Bioinformatics. 2016. PMID: 26424855 - A comprehensive overview of Infinium HumanMethylation450 data processing.
Dedeurwaerder S, Defrance M, Bizet M, Calonne E, Bontempi G, Fuks F. Dedeurwaerder S, et al. Brief Bioinform. 2014 Nov;15(6):929-41. doi: 10.1093/bib/bbt054. Epub 2013 Aug 29. Brief Bioinform. 2014. PMID: 23990268 Free PMC article. Review. - Base resolution methylome profiling: considerations in platform selection, data preprocessing and analysis.
Sun Z, Cunningham J, Slager S, Kocher JP. Sun Z, et al. Epigenomics. 2015 Aug;7(5):813-28. doi: 10.2217/epi.15.21. Epub 2015 Sep 14. Epigenomics. 2015. PMID: 26366945 Free PMC article. Review.
Cited by
- Molecular and cell phenotype programs in oral epithelial cells directed by co-exposure to arsenic and smokeless tobacco.
Das S, Thakur S, Cahais V, Virard F, Claeys L, Renard C, Cuenin C, Cros MP, Keïta S, Venuti A, Sirand C, Ghantous A, Herceg Z, Korenjak M, Zavadil J. Das S, et al. bioRxiv [Preprint]. 2024 Oct 15:2024.10.14.618077. doi: 10.1101/2024.10.14.618077. bioRxiv. 2024. PMID: 39463997 Free PMC article. Preprint. - A novel DNA methylation-based surrogate biomarker for chronic systemic inflammation (InfLaMeS): results from the Health and Retirement Study.
Meier HCS, Klopack ET, Farnia MP, Hernandez B, Mitchell C, Faul JD, McCrory C, Kenny RA, Crimmins EM. Meier HCS, et al. medRxiv [Preprint]. 2024 Oct 15:2024.10.11.24315339. doi: 10.1101/2024.10.11.24315339. medRxiv. 2024. PMID: 39484273 Free PMC article. Preprint. - Genome-wide DNA methylation analysis pre- and post-lenalidomide treatment in patients with myelodysplastic syndrome with isolated deletion (5q).
Hecht A, Meyer JA, Jann JC, Sockel K, Giagounidis A, Götze KS, Letsch A, Haase D, Schlenk RF, Haferlach T, Schafhausen P, Bug G, Lübbert M, Thol F, Büsche G, Schuler E, Nowak V, Obländer J, Fey S, Müller N, Metzgeroth G, Hofmann WK, Germing U, Nolte F, Reinwald M, Nowak D. Hecht A, et al. Ann Hematol. 2021 Jun;100(6):1463-1471. doi: 10.1007/s00277-021-04492-1. Epub 2021 Apr 27. Ann Hematol. 2021. PMID: 33903952 Free PMC article. Clinical Trial. - DNA methylation signatures in cord blood associated with birthweight are enriched for dmCpGs previously associated with maternal hypertension or pre-eclampsia, smoking and folic acid intake.
Antoun E, Titcombe P, Dalrymple K, Kitaba NT, Barton SJ, Flynn A, Murray R, Garratt ES, Seed PT, White SL, Cooper C, Inskip HM, Hanson M, Poston L, Godfrey KM, Lillycrop KA; UPBEAT Consortium/EpiGen Consortium. Antoun E, et al. Epigenetics. 2022 Apr;17(4):405-421. doi: 10.1080/15592294.2021.1908706. Epub 2021 Apr 28. Epigenetics. 2022. PMID: 33784941 Free PMC article. - Analysis of Systemic Epigenetic Alterations in Inflammatory Bowel Disease: Defining Geographical, Genetic and Immune-Inflammatory influences on the Circulating Methylome.
Kalla R, Adams AT, Nowak JK, Bergemalm D, Vatn S, Ventham NT, Kennedy NA, Ricanek P, Lindstrom J; IBD-Character Consortium; Söderholm J, Pierik M, D'Amato M, Gomollón F, Olbjørn C, Richmond R, Relton C, Jahnsen J, Vatn MH, Halfvarson J, Satsangi J. Kalla R, et al. J Crohns Colitis. 2023 Mar 18;17(2):170-184. doi: 10.1093/ecco-jcc/jjac127. J Crohns Colitis. 2023. PMID: 36029471 Free PMC article.
References
- Bibikova M, et al. High density DNA methylation array with single CpG site resolution. Genomics. 2011;98:288–295. - PubMed
- Bolstad BM, et al. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. - PubMed
- Chambers JM. Programming with Data: A Guide to the S Language. New York: Springer; 1998.
Publication types
MeSH terms
Grants and funding
- P50 HG003233/HG/NHGRI NIH HHS/United States
- R01 GM083084/GM/NIGMS NIH HHS/United States
- R01 AG042187/AG/NIA NIH HHS/United States
- R01AG042187/AG/NIA NIH HHS/United States
- P50HG003233/HG/NHGRI NIH HHS/United States
- R01 GM103552/GM/NIGMS NIH HHS/United States
- GM083084/GM/NIGMS NIH HHS/United States
- GM103552/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources