Role of phosphatidylserine receptors in enveloped virus infection - PubMed (original) (raw)
Role of phosphatidylserine receptors in enveloped virus infection
Kouki Morizono et al. J Virol. 2014 Apr.
Abstract
We recently demonstrated that a soluble protein, Gas6, can facilitate viral entry by bridging viral envelope phosphatidylserine to Axl, a receptor tyrosine kinase expressed on target cells. The interaction between phosphatidylserine, Gas6, and Axl was originally shown to be a molecular mechanism through which phagocytes recognize phosphatidylserine exposed on dead cells. Since our initial report, several groups have confirmed that Axl/Gas6, as well as other phosphatidylserine receptors, facilitate entry of dengue, West Nile, and Ebola viruses. Virus binding by viral envelope phosphatidylserine is now a viral entry mechanism generalized to many families of viruses. In addition to Axl/Gas6, various molecules are known to recognize phosphatidylserine; however, the effects of these molecules on virus binding and entry have not been comprehensively evaluated and compared. In this study, we examined most of the known human phosphatidylserine-recognizing molecules, including MFG-E8, TIM-1, -3, and -4, CD300a, BAI1, and stabilin-1 and -2, for their abilities to facilitate virus binding and infection. Using pseudotyped lentiviral vectors, we found that a soluble phosphatidylserine-binding protein, MFG-E8, enhances transduction. Cell surface receptors TIM-1 and -4 also enhance virus binding/transduction. The extent of enhancement by these molecules varies, depending on the type of pseudotyping envelope proteins. Mutated MFG-E8, which binds viral envelope phosphatidylserine without bridging virus to cells, but, surprisingly, not annexin V, which has been used to block phagocytosis of dead cells by concealing phosphatidylserine, efficiently blocks these phosphatidylserine-dependent viral entry mechanisms. These results provide insight into understanding the role of viral envelope phosphatidylserine in viral infection.
Importance: Envelope phosphatidylserine has previously been shown to be important for replication of various envelope viruses, but details of this mechanism(s) were unclear. We were the first to report that a bifunctional serum protein, Gas6, bridges envelope phosphatidylserine to a cell surface receptor, Axl. Recent studies demonstrated that many envelope viruses, including vaccinia, dengue, West Nile, and Ebola viruses, utilize Axl/Gas6 to facilitate their entry, suggesting that the phosphatidylserine-mediated viral entry mechanism can be shared by various enveloped viruses. In addition to Axl/Gas6, various molecules are known to recognize phosphatidylserine; however, the effects of these molecules on virus binding and entry have not been comprehensively evaluated and compared. In this study, we examined most human phosphatidylserine-recognizing molecules for their abilities to facilitate viral infection. The results provide insights into the role(s) of envelope phosphatidylserine in viral infection, which can be applicable to the development of novel antiviral reagents that block phosphatidylserine-mediated viral entry.
Figures
FIG 1
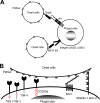
Molecular mechanisms of phagocytosis of dead cells. (A) Molecular mechanisms of phagocytosis of dead cells mediated by soluble molecules that bridge the PtdSer exposed on dead cells to receptors on phagocytes. (B) Molecular mechanisms of phagocytosis of dead cells mediated by receptors that directly recognize PtdSer. TIM-1, -3, and -4 and CD300a bind PtdSer via their N-terminal IgV domains. TIM-1 and -4 each have one cytoplasmic tyrosine residue and CD300a has four cytoplasmic tyrosine residues; these residues are phosphorylated and mediate signaling after binding to PtdSer (red circles). BAI1 is a seven-transmembrane receptor. Its extracellular N terminus has five thrombospondin type 1 repeat domains that bind PtdSer. The N termini of stabilin-1 and -2 each have four clusters of epidermal growth factor-like domain repeats that bind PtdSer. The cytoplasmic domains of BAI1 and stabilin-1 and -2 can activate Rac1 GTPase. In addition to these receptors, RAGE has recently been identified to bind PtdSer; however, its PtdSer-binding domain(s) has not yet been identified.
FIG 2
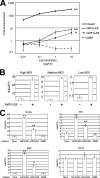
Binding of lentiviral vectors mediated by soluble PtdSer-recognizing molecules. (A) Schematic representation of chimeric Sindbis virus Envs. The Sindbis virus Env is first synthesized as a polypeptide and subsequently cleaved by cellular proteases to generate E3, E2, 6K, and E1 proteins. E1 and E2 are incorporated into the viral envelope. Both the 2.2 and 2.2 1L1L pseudotypes are derived from the wild-type Sindbis virus Env with four mutations (red lines) to reduce their binding to original receptors while maintaining fusion activity. In addition, 2.2 1L1L has two flexible linkers (Gly-Gly-Gly-Gly-Ser) at amino acid (a.a.) 70 of the E2 protein, and 2.2 contains the IgG-binding domain of protein A (ZZ), which was inserted into the E2 region at amino acid 70. (B) Expression levels of Axl (red line), integrin αVβ3 (blue line), and integrin αVβ5 (green line) were analyzed by flow cytometry after staining with antibodies specific to each antigen. The isotype of all antibodies was IgG1; thus, an IgG1 isotype control antibody was used for negative-control staining (black line). (C) Schematic representation of recombinant hGas6, hMFG-E8, mMFG-E8, and D89E, a mutant of MFG-E8 that cannot bind integrins. Gla, γ-carboxyglutamic acid domain; EGF, E1, and E2, epidermal growth factor-like domains; SHBG, sex hormone-binding globulin domain; C1 and C2, discoid domains; P/T, proline/threonine-enriched motif. The γ-carboxyglutamic acid domain of Gas6 and the C2 domain of MFG-E8 bind PtdSer. (D) Staining of HMVECs with an APC-conjugated anti-Flag tag antibody after incubation with hGas6 (red line), mMFG-E8 (blue line), or D89E (green line). The black line represents staining without prior incubation with any ligand. (E) Binding of the EGFP-labeled 2.2 1L1L pseudotype to HMVECs after incubation with hGas6, hMFG-E8, or mMFG-E8. Black line, HMVECs without virus; red line, HMVECs with virus with or without the bridging molecules indicated in the figure. The MFI values correspond to those for EGFP signals, which increase upon binding of EGFP-labeled virus. This experiment was repeated twice in singlicate and once in triplicate, and the MFI values shown are the averages and standard deviations of the triplicate experiment; representative flow cytometry histograms are shown. Significance was calculated by comparing the value of virus binding without a bridging molecule to the value of virus binding in the presence of the bridging molecules indicated in the figure using a two-sample Student t test (**, P < 0.01).
FIG 3
Lentiviral transduction mediated by MFG-E8. (A) Enhancement of HMVEC transduction with the 2.2 1L1L pseudotype (100 ng p24/ml) after incubation with hGas6, hMFG-E8, mMFG-E8, or D89E at the concentrations shown on the x axis. The fold enhancement was calculated by the following formula: percentage of EGFP-expressing cells preincubated with each ligand divided by percentage of EGFP-expressing cells preincubated with buffer only. This experiment was repeated twice in singlicate and twice in triplicate, and the results shown are the averages and standard deviations of one representative triplicate experiment. The proportion of EGFP-expressing cells preincubated with buffer only was 0.12% ± 0.05% (average ± standard deviation). Significance was calculated by comparing the value for the no-protein samples to the value for the samples with 10 μg/ml proteins, using a two-sample Student t test (*, P < 0.05; **, P < 0.01). (B) Transduction of HMVECs with the 2.2 1L1L pseudotype with or without preincubation with hMFG-E8 at high (2,500 ng p24/ml), medium (500 ng p24/ml), and low (100 ng p24/ml) MOIs. This experiment was repeated twice in triplicate, and the results shown are the averages and standard deviations of one representative triplicate experiment. (C) Transduction of HMVECs with lentiviral vectors pseudotyped with Sindbis (100 ng p24/ml), RRV (400 ng p24/ml), gp64 (400 ng p24/ml), or VSV-G (8 ng p24/ml) after preincubation with hMFG-E8, mMFG-E8, or D89E at 3 μg/ml. This experiment was repeated once in singlicate and twice in triplicate, and the results shown are the averages and standard deviations of one representative triplicate experiment. Significance was calculated by comparing the value of transduction obtained with the no-protein samples to the value of transduction obtained with the proteins indicated in the figure using a two-sample Student t test (*, P < 0.05; **, P < 0.01).
FIG 4
Role of viral envelope PtdSer in MFG-E8-mediated lentiviral transduction. (A) Blocking of MFG-E8-mediated transduction of HMVECs with the 2.2 1L1L pseudotype by ANX V or D89E. HMVECs were incubated with hMFG-E8 (3 μg/ml) for 4 h prior to transduction. The 2.2 1L1L pseudotype (500 ng p24/ml) was preincubated with ANX V or D89E for 1 h at the concentrations shown in the figure prior to transduction. HMVECs were incubated for 2 h with the 2.2 1L1L pseudotype in the presence of ANX V or D89E. This experiment was repeated twice in singlicate and twice in triplicate, and the results shown are the averages and standard deviations of one representative triplicate experiment. (B) Blocking of MFG-E8-mediated transduction of HMVECs with the 2.2 1L1L pseudotype (100 ng p24/ml) by liposomes. HMVECs were incubated with hMFG-E8 (3 μg/ml) for 4 h, followed by incubation for 1 h with liposomes consisting of either PtdChl only (PtdChl), 75% PtdChl and 25% PtdSer (PtdSer), or 75% PtdChl and 25% PtdEtr (PtdEtr). The cells were then transduced with the 2.2 1L1L pseudotype for 2 h in the presence of the same liposomes. This experiment was repeated twice in singlicate and twice in triplicate, and the results shown are the averages and standard deviations of one representative triplicate experiment. For panels A and B, significance was calculated using a two-sample Student t test by comparing the MFG-E8-mediated transduction without blocking reagents to that with the blocking reagents indicated in the figure (*, P < 0.05; **, P < 0.01).
FIG 5
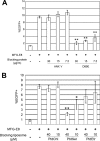
Ectopic expression of PtdSer receptors on 293T cells. (A) Schematic representation of lentiviral vectors expressing TIM-1, -3, and -4, CD300a, or BAI1 and plasmid vectors expressing stabilin-1 and -2. SIN LTR, self-inactivating long terminal repeat; Ubc pro, ubiquitin C promoter; SV40 pro, SV40 promoter; TK, thymidine kinase. (B) Expression of Axl, TIM-1, -3, and -4, CD300a, BAI1, and stabilin-1 and -2 on 293T cells ectopically expressing each of these receptors analyzed by the use of antibodies specific to each receptor. Black line, staining by control antibodies; red line, staining by antibodies specific to each PtdSer receptors. (C) (Left column and top four panels of right column) Expression of TIM-1, -3, and -4, CD300a, BAI1, and stabilin-1 and -2 on 293T cells ectopically expressing each of these receptors and parental 293T cells analyzed by the antibody specific to the Flag tag. Black line, staining by APC-conjugated isotype control antibody; red line, staining by APC-conjugated anti-Flag tag antibody. (Bottom two panels of right column) Expression of Axl was analyzed by incubating 293T and Axl 293T cells with hGas6, followed by staining with the anti-Flag tag antibody. Black line, staining by APC-conjugated anti-Flag tag antibody without preincubation with hGas6; red line, staining by APC-conjugated anti-Flag tag antibody after incubation with hGas6.
FIG 6
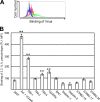
Binding of the 2.2 1L1L pseudotype mediated by PtdSer receptors. (A) Representative flow cytometry analysis data for the binding of the EGFP-labeled 2.2 1L1L pseudotype (100 ng p24) to 293T cells and to Axl 293T cells preincubated with hGas6. Black line, 293T cells without virus; red line, 293T cells with virus; blue line, Axl 293T cells preincubated with hGas6 without virus; green line, Axl 293T cells preincubated with hGas6 with virus. (B) Quantitation of EGFP-labeled 2.2 1L1L pseudotype binding to 293T cells, Axl 293T cells preincubated with hGas6, and TIM-1, -3, or -4, CD300a, BAI1, stabilin-1 or -2, STAB 2-15, STAB 2-17, and STAB 2-20 293T cells. The values on the y axis correspond to the MFIs of the EGFP signals, which increase by binding of EGFP-labeled virus. This experiment was repeated once in singlicate and twice in triplicate, and the results shown are the averages and standard deviations of one representative triplicate experiment. Significance was calculated by comparing the value for 293T cells to the values for the cells expressing each type of PtdSer receptor using a two-sample Student t test (**, P < 0.01).
FIG 7
Lentiviral transduction mediated by PtdSer receptors. Percentage of EGFP-expressing 293T cells; Axl 293T cells preincubated with hGas6, and TIM-1, -3, or -4, CD300a, BAI1, stabilin-1 or -2, STAB 2-15, STAB 2-17, or STAB 2-20 293T cells after transduction with lentiviral vectors pseudotyped with 2.2 1L1L (50 ng p24/ml) (A), Sindbis (20 ng p24/ml) (B), RRV (40 ng p24/ml) (C), gp64 (25 ng p24/ml) (D), or VSV-G (10 ng p24/ml) (E). EGFP expression was analyzed at 3 days postransduction. The results are the averages of triplicate experiments and are shown with standard deviations. (F) Percentage of EGFP-expressing 293T cells and Flag− TIM-1, -3, or -4, Flag− CD300a, Flag− BAI1, Flag− stabilin-1 or -2, STAB 2-5, and STAB 2-8 293T cells after transduction with the 2.2 1L1L pseudotype (50 ng p24/ml). (G) Titers (EGFP transduction units [TU]/ml) of various pseudotypes with 293T cells, Axl 293T cells preincubated with hGas6, and TIM-1 293T cells as target cells. The titers came from concentrated virus stocks that contained 60 μg p24/ml. The experiments whose results are presented in panels A and F were repeated three times in singlicate and twice in triplicate, and the results shown are the averages and standard deviations of one representative triplicate experiment. The experiments whose results are presented in panels B to E were repeated twice in triplicate, and the results shown are the averages and standard deviations of one representative triplicate experiment. For panels A to F, significance was calculated by comparing the transduction efficiencies of 293T cells to those of PtdSer receptor-expressing cells using a two-sample Student t test (*, P < 0.05; **, P < 0.01).
FIG 8
Role of viral envelope PtdSer in TIM-1- and TIM-4-mediated lentiviral transduction. (A) Blocking of TIM-1-mediated 2.2 1L1L pseudotype transduction by ANX V or D89E. The 2.2 1L1L pseudotype (50 ng p24/ml) was incubated with ANX V or D89E for 1 h prior to transduction at the concentrations indicated in the figure. The vectors were then used for transduction of 293T and TIM-1 293T cells. (B) Blocking of TIM-4-mediated gp64 pseudotype transduction by ANX V or D89E. The gp64 pseudotype (25 ng p24/ml) was incubated with ANX V or D89E for 1 h prior to transduction at the concentrations indicated in the figure. The vectors were then used for the transduction of 293T and TIM-4 293T cells. (C) Blocking of TIM-1-mediated 2.2 1L1L pseudotype transduction by liposomes. TIM-1 293T cells were incubated for 1 h with the liposomes indicated in the figure. The cells were then transduced with the 2.2 1L1L pseudotype (50 ng p24/ml) for 2 h in the presence of the same liposomes. (D) Blocking of TIM-4-mediated gp64 pseudotype transduction by liposomes. TIM-4 293T cells were incubated for 1 h with the liposomes indicated in the figure. The cells were then transduced with the gp64 pseudotype (25 ng p24/ml) for 2 h in the presence of the same liposomes. (E) Blocking by D89E of HLA class I-mediated transduction of 293T cells with the 2.2 pseudotype. The 2.2 pseudotype (100 ng p24/ml) was conjugated with an anti-HLA class I antibody (Ab), followed by 1 h of incubation with D89E at the concentrations indicated in the figure. The vectors were then used for transduction of 293T cells. The experiments whose results are presented in panels A, B, and E were repeated twice in singlicate and twice in triplicate, and the results shown are the averages and standard deviations of one representative triplicate experiment. The experiments whose results are presented in panels C and D were repeated twice in triplicate, and the results shown are the averages and standard deviations of one representative triplicate experiment. For panels A to D, significance was calculated by comparing the results of TIM-1- or TIM-4-mediated transduction without blocking reagents to those with blocking reagents using a two-sample Student t test (*, P < 0.05; **, P < 0.01).
Similar articles
- Phosphatidylethanolamine and Phosphatidylserine Synergize To Enhance GAS6/AXL-Mediated Virus Infection and Efferocytosis.
Zhang L, Richard AS, Jackson CB, Ojha A, Choe H. Zhang L, et al. J Virol. 2020 Dec 22;95(2):e02079-20. doi: 10.1128/JVI.02079-20. Print 2020 Dec 22. J Virol. 2020. PMID: 33115868 Free PMC article. - Characterizing functional domains for TIM-mediated enveloped virus entry.
Moller-Tank S, Albritton LM, Rennert PD, Maury W. Moller-Tank S, et al. J Virol. 2014 Jun;88(12):6702-13. doi: 10.1128/JVI.00300-14. Epub 2014 Apr 2. J Virol. 2014. PMID: 24696470 Free PMC article. - The soluble serum protein Gas6 bridges virion envelope phosphatidylserine to the TAM receptor tyrosine kinase Axl to mediate viral entry.
Morizono K, Xie Y, Olafsen T, Lee B, Dasgupta A, Wu AM, Chen IS. Morizono K, et al. Cell Host Microbe. 2011 Apr 21;9(4):286-98. doi: 10.1016/j.chom.2011.03.012. Cell Host Microbe. 2011. PMID: 21501828 Free PMC article. - Phosphatidylserine receptors: enhancers of enveloped virus entry and infection.
Moller-Tank S, Maury W. Moller-Tank S, et al. Virology. 2014 Nov;468-470:565-580. doi: 10.1016/j.virol.2014.09.009. Epub 2014 Sep 29. Virology. 2014. PMID: 25277499 Free PMC article. Review. - Roles of phosphatidylserine exposed on the viral envelope and cell membrane in HIV-1 replication.
Chua BA, Ngo JA, Situ K, Morizono K. Chua BA, et al. Cell Commun Signal. 2019 Oct 21;17(1):132. doi: 10.1186/s12964-019-0452-1. Cell Commun Signal. 2019. PMID: 31638994 Free PMC article. Review.
Cited by
- Phosphatidylserine-mediated uptake of extracellular vesicles by hepatocytes ameliorates liver ischemia-reperfusion injury.
Li R, Wang C, Chen X, Fu E, Zhang K, Tao H, Han Z, Han ZC, Li Z. Li R, et al. Apoptosis. 2024 Oct 13. doi: 10.1007/s10495-024-02030-8. Online ahead of print. Apoptosis. 2024. PMID: 39397123 - Insights into mosquito-borne arbovirus receptors.
Liu J, Quan Y, Tong H, Zhu Y, Shi X, Liu Y, Cheng G. Liu J, et al. Cell Insight. 2024 Aug 24;3(6):100196. doi: 10.1016/j.cellin.2024.100196. eCollection 2024 Dec. Cell Insight. 2024. PMID: 39391003 Free PMC article. Review. - Could Gas6/TAM Axis Provide Valuable Insights into the Pathogenesis of Systemic Sclerosis?
Apostolo D, D'Onghia D, Nerviani A, Ghirardi GM, Sola D, Perazzi M, Tonello S, Colangelo D, Sainaghi PP, Bellan M. Apostolo D, et al. Curr Issues Mol Biol. 2024 Jul 15;46(7):7486-7504. doi: 10.3390/cimb46070444. Curr Issues Mol Biol. 2024. PMID: 39057085 Free PMC article. Review. - Gas6/TAM system as potential biomarker for multiple sclerosis prognosis.
D'Onghia D, Colangelo D, Bellan M, Tonello S, Puricelli C, Virgilio E, Apostolo D, Minisini R, Ferreira LL, Sozzi L, Vincenzi F, Cantello R, Comi C, Pirisi M, Vecchio D, Sainaghi PP. D'Onghia D, et al. Front Immunol. 2024 Apr 30;15:1362960. doi: 10.3389/fimmu.2024.1362960. eCollection 2024. Front Immunol. 2024. PMID: 38745659 Free PMC article. - Envelope protein-specific B cell receptors direct lentiviral vector tropism in vivo.
Takano KA, Wong AAL, Brown R, Situ K, Chua BA, Abu AE, Pham TT, Reyes GC, Ramachandran S, Kamata M, Li MMH, Wu TT, Rao DS, Arumugaswami V, Dorshkind K, Cole S, Morizono K. Takano KA, et al. Mol Ther. 2024 May 1;32(5):1311-1327. doi: 10.1016/j.ymthe.2024.03.002. Epub 2024 Mar 6. Mol Ther. 2024. PMID: 38449314
References
- Callahan MK, Popernack PM, Tsutsui S, Truong L, Schlegel RA, Henderson AJ. 2003. Phosphatidylserine on HIV envelope is a cofactor for infection of monocytic cells. J. Immunol. 170:4840–4845 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI108400/AI/NIAID NIH HHS/United States
- UL1TR000124/TR/NCATS NIH HHS/United States
- R21AI095004/AI/NIAID NIH HHS/United States
- AI28697/AI/NIAID NIH HHS/United States
- UL1 TR000124/TR/NCATS NIH HHS/United States
- R21 AI095004/AI/NIAID NIH HHS/United States
- R01AI108400/AI/NIAID NIH HHS/United States
- P30 CA016042/CA/NCI NIH HHS/United States
- P30 AI028697/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous