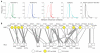Exome sequencing links corticospinal motor neuron disease to common neurodegenerative disorders - PubMed (original) (raw)
. 2014 Jan 31;343(6170):506-511.
doi: 10.1126/science.1247363.
Ali G Fenstermaker # 1, Maha S Zaki # 2, Matan Hofree 3, Jennifer L Silhavy 1, Andrew D Heiberg 1, Mostafa Abdellateef 1, Basak Rosti 1, Eric Scott 1, Lobna Mansour 4, Amira Masri 5, Hulya Kayserili 6, Jumana Y Al-Aama 7, Ghada M H Abdel-Salam 2, Ariana Karminejad 8, Majdi Kara 9, Bulent Kara 10, Bita Bozorgmehri 8, Tawfeg Ben-Omran 11, Faezeh Mojahedi 12, Iman Gamal El Din Mahmoud 4, Naima Bouslam 13, Ahmed Bouhouche 13, Ali Benomar 13, Sylvain Hanein 14, Laure Raymond 14, Sylvie Forlani 14, Massimo Mascaro 1, Laila Selim 4, Nabil Shehata 15, Nasir Al-Allawi 16, P S Bindu 17, Matloob Azam 18, Murat Gunel 19, Ahmet Caglayan 19, Kaya Bilguvar 19, Aslihan Tolun 20, Mahmoud Y Issa 2, Jana Schroth 1, Emily G Spencer 1, Rasim O Rosti 1, Naiara Akizu 1, Keith K Vaux 1, Anide Johansen 1, Alice A Koh 1, Hisham Megahed 2, Alexandra Durr 14 21, Alexis Brice 14 21 22, Giovanni Stevanin 14 21 22 23, Stacy B Gabriel 24, Trey Ideker 3, Joseph G Gleeson 1
Affiliations
- PMID: 24482476
- PMCID: PMC4157572
- DOI: 10.1126/science.1247363
Exome sequencing links corticospinal motor neuron disease to common neurodegenerative disorders
Gaia Novarino et al. Science. 2014.
Abstract
Hereditary spastic paraplegias (HSPs) are neurodegenerative motor neuron diseases characterized by progressive age-dependent loss of corticospinal motor tract function. Although the genetic basis is partly understood, only a fraction of cases can receive a genetic diagnosis, and a global view of HSP is lacking. By using whole-exome sequencing in combination with network analysis, we identified 18 previously unknown putative HSP genes and validated nearly all of these genes functionally or genetically. The pathways highlighted by these mutations link HSP to cellular transport, nucleotide metabolism, and synapse and axon development. Network analysis revealed a host of further candidate genes, of which three were mutated in our cohort. Our analysis links HSP to other neurodegenerative disorders and can facilitate gene discovery and mechanistic understanding of disease.
Figures
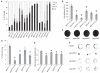
Fig. 1. Functional validation of private HSP genes in zebrafish
(A) Quantification of 24-hours-post-fertilization (hpf) embryos mortality (black) and curly-tail (gray) phenotypes for noninjected (NI), scrambled, and morphants (MO) at stated nanogram concentrations. Overt phenotypes were observed for all MOs except MOpgap1. (B) Average touch-response distance (in arbitrary units, A.U.) in 72-hpf larvae, showing blunted response for all MOs. (C) Immediate touch-response trajectory of example larvae, each shaded uniquely. Mars2 MO was too severe to be tested, whereas others showed reduced response. (D to F) Spontaneous locomotion at 6 days post fertilization. (D) Average percent of time spent moving over a 30-min window showed a reduction for all for at least one dose. (E) Average active period duration, showing reduction for all. (F) Representative kymographs recording fish position (black dot) over 30-min recording. MOs showed either short distance traveled (MOarl6ip1) or reduced movements per recording (MOpgap1 and MOusp8). *P < 0.01 (t test). _N_ > 2 experiments with n > 20 animals per experiment. Error bars indicate standard error.
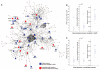
Fig. 2. Hereditary spastic paraplegia interactome
(A) HSP seeds + candidate network (edge-weighted force-directed layout), demonstrating many of the genes known to be mutated in HSP (seeds, blue) and new HSP candidates (red), along with others (circles) constituting the network. (B and C) Comparison of statistical strength of HSP subnetworks with 10,000 permutations of randomly selected proteins. Dots denote the value of the metric on the true set (i.e., seeds or seeds + candidates). Box and whisker plots denote matched null distributions (i.e., 10,000 permutations). (B) Seed (known mutated in HSP) versus random proteins drawn with the same degree distribution. (C) Seed plus candidate HSP versus a matching set of proteins. (Left) Within group edge count (i.e., number of edges between members of the query set). (Middle) Interaction neighborhood overlap (i.e., Jaccard similarity). (Right) Network random walk similarity.
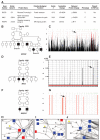
Fig. 3. Genes from HSP networks found mutated in HSP
(A) HSP candidate genes predicted from the HSPome found mutated in the HSP cohort. BICD2, MAP, and REEP2 were subsequently found mutated in HSP families 1370 (B), 1226 (D), and 1967 (F), respectively. (C) Homozygosity plot from family 1370. Red bars, regions of homozygosity; arrow, homozygous block containing BICD2. (E) Linkage plot of family 1226; arrow, MAG locus. (G) Homozygosity plot; arrow, REEP2 locus. (H to J) Zoom in from HSPome for specific interaction identifying candidates CCDC64 (a paralog of BIC2D), MAG, and REEP2 (yellow) with previously published (blue) and newly identified (red) genes mutated in HSP. Blue lines denote manually curated interactions.
Fig. 4. Functional link between HSP genes and genes of other neurodegenerative conditions
(A) Density distribution representing random walk distances of OMIM-derived neurodegeneration gene networks along with 10,000 permutations of randomly selected protein pools compared with the HSP seeds plus candidates pool. The top 5%ile distance is shaded. Only for Parkinson’s, Alzheimer’s, and ALS do the HSP seeds plus candidates fall within this 5%, whereas epilepsy and autism spectrum disorder show no statistical overlap. (B) Bipartite network showing the top links between the set of HSP and ALS proteins. Clear circles, HSP seeds; yellow circles, HSP candidates; boxes, ALS genes (VCP and ALS2 are implicated as causative of both HSP and ALS); line thickness, diffusion similarity between the two proteins.
Comment in
- Genetics. A unified process for neurological disease.
Singleton AB. Singleton AB. Science. 2014 Jan 31;343(6170):497-8. doi: 10.1126/science.1250172. Science. 2014. PMID: 24482474 Free PMC article. - Genetics: Exome sequencing sheds light on hereditary spastic paraplegia.
Chase A. Chase A. Nat Rev Neurol. 2014 Mar;10(3):124. doi: 10.1038/nrneurol.2014.27. Epub 2014 Feb 25. Nat Rev Neurol. 2014. PMID: 24566998 No abstract available. - Will next-generation sequencing also take us to the next level of understanding disease mechanisms?
Lehn A. Lehn A. Mov Disord. 2014 Jun;29(7):868. doi: 10.1002/mds.25905. Epub 2014 May 5. Mov Disord. 2014. PMID: 24797999 No abstract available.
Similar articles
- Genetics: Exome sequencing sheds light on hereditary spastic paraplegia.
Chase A. Chase A. Nat Rev Neurol. 2014 Mar;10(3):124. doi: 10.1038/nrneurol.2014.27. Epub 2014 Feb 25. Nat Rev Neurol. 2014. PMID: 24566998 No abstract available. - Will next-generation sequencing also take us to the next level of understanding disease mechanisms?
Lehn A. Lehn A. Mov Disord. 2014 Jun;29(7):868. doi: 10.1002/mds.25905. Epub 2014 May 5. Mov Disord. 2014. PMID: 24797999 No abstract available. - Genetics. A unified process for neurological disease.
Singleton AB. Singleton AB. Science. 2014 Jan 31;343(6170):497-8. doi: 10.1126/science.1250172. Science. 2014. PMID: 24482474 Free PMC article. - Hereditary spastic paraplegia: More than an upper motor neuron disease.
Parodi L, Fenu S, Stevanin G, Durr A. Parodi L, et al. Rev Neurol (Paris). 2017 May;173(5):352-360. doi: 10.1016/j.neurol.2017.03.034. Epub 2017 Apr 24. Rev Neurol (Paris). 2017. PMID: 28449883 Review. - Hereditary spastic paraplegia: Genetic heterogeneity and common pathways.
Panza E, Meyyazhagan A, Orlacchio A. Panza E, et al. Exp Neurol. 2022 Nov;357:114203. doi: 10.1016/j.expneurol.2022.114203. Epub 2022 Aug 13. Exp Neurol. 2022. PMID: 35970204 Review.
Cited by
- Hereditary spastic paraplegia SPG4: what is known and not known about the disease.
Solowska JM, Baas PW. Solowska JM, et al. Brain. 2015 Sep;138(Pt 9):2471-84. doi: 10.1093/brain/awv178. Epub 2015 Jun 20. Brain. 2015. PMID: 26094131 Free PMC article. Review. - Genotyping KIF1C (c.608G>A) Mutant Reveals a Wide Distribution of Progressive Ataxia in German Charolais Cattle.
Bischofberger FM, Reinartz S, Distl O. Bischofberger FM, et al. Animals (Basel). 2024 Jan 23;14(3):366. doi: 10.3390/ani14030366. Animals (Basel). 2024. PMID: 38338009 Free PMC article. - Alternative splicing during mammalian organ development.
Mazin PV, Khaitovich P, Cardoso-Moreira M, Kaessmann H. Mazin PV, et al. Nat Genet. 2021 Jun;53(6):925-934. doi: 10.1038/s41588-021-00851-w. Epub 2021 May 3. Nat Genet. 2021. PMID: 33941934 Free PMC article. - Drosophila and experimental neurology in the post-genomic era.
Shulman JM. Shulman JM. Exp Neurol. 2015 Dec;274(Pt A):4-13. doi: 10.1016/j.expneurol.2015.03.016. Epub 2015 Mar 24. Exp Neurol. 2015. PMID: 25814441 Free PMC article. Review. - Hook3 is a scaffold for the opposite-polarity microtubule-based motors cytoplasmic dynein-1 and KIF1C.
Kendrick AA, Dickey AM, Redwine WB, Tran PT, Vaites LP, Dzieciatkowska M, Harper JW, Reck-Peterson SL. Kendrick AA, et al. J Cell Biol. 2019 Sep 2;218(9):2982-3001. doi: 10.1083/jcb.201812170. Epub 2019 Jul 18. J Cell Biol. 2019. PMID: 31320392 Free PMC article.
References
- See supplementary text available on Science Online.
- D’Amico A, et al. Neurology. 2004;62:2138–2139. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P41 GM103504/GM/NIGMS NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- R01 NS048453/NS/NINDS NIH HHS/United States
- N01 CO012400/CA/NCI NIH HHS/United States
- P01HD070494/HD/NICHD NIH HHS/United States
- U54 HG003067/HG/NHGRI NIH HHS/United States
- P30NS047101/NS/NINDS NIH HHS/United States
- R01NS048453/NS/NINDS NIH HHS/United States
- P30 NS047101/NS/NINDS NIH HHS/United States
- U54HG003067/HG/NHGRI NIH HHS/United States
- R01 NS041537/NS/NINDS NIH HHS/United States
- U54HG006504/HG/NHGRI NIH HHS/United States
- U54 HG006504/HG/NHGRI NIH HHS/United States
- R01 NS052455/NS/NINDS NIH HHS/United States
- P01 HD070494/HD/NICHD NIH HHS/United States
- N01-CO-12400/CO/NCI NIH HHS/United States
- HHSN268201100011/PHS HHS/United States
- R01NS041537/NS/NINDS NIH HHS/United States
- R01NS052455/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases