voom: Precision weights unlock linear model analysis tools for RNA-seq read counts - PubMed (original) (raw)
voom: Precision weights unlock linear model analysis tools for RNA-seq read counts
Charity W Law et al. Genome Biol. 2014.
Abstract
New normal linear modeling strategies are presented for analyzing read counts from RNA-seq experiments. The voom method estimates the mean-variance relationship of the log-counts, generates a precision weight for each observation and enters these into the limma empirical Bayes analysis pipeline. This opens access for RNA-seq analysts to a large body of methodology developed for microarrays. Simulation studies show that voom performs as well or better than count-based RNA-seq methods even when the data are generated according to the assumptions of the earlier methods. Two case studies illustrate the use of linear modeling and gene set testing methods.
Figures
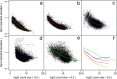
Figure 1
Mean-variance relationships. Gene-wise means and variances of RNA-seq data are represented by black points with a LOWESS trend. Plots are ordered by increasing levels of biological variation in datasets. (a) voom trend for HBRR and UHRR genes for Samples A, B, C and D of the SEQC project; technical variation only. (b) C57BL/6J and DBA mouse experiment; low-level biological variation. (c) Simulation study in the presence of 100 upregulating genes and 100 downregulating genes; moderate-level biological variation. (d) Nigerian lymphoblastoid cell lines; high-level biological variation. (e)Drosophila melanogaster embryonic developmental stages; very high biological variation due to systematic differences between samples. (f) LOWESS voom trends for datasets (a)–(e). HBRR, Ambion’s Human Brain Reference RNA; LOWESS, locally weighted regression; UHRR, Stratagene’s Universal Human Reference RNA.
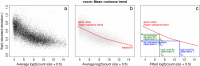
Figure 2
voom mean-variance modeling. (a) Gene-wise square-root residual standard deviations are plotted against average log-count. (b) A functional relation between gene-wise means and variances is given by a robust LOWESS fit to the points. (c) The mean-variance trend enables each observation to map to a square-root standard deviation value using its fitted value for log-count. LOWESS, locally weighted regression.
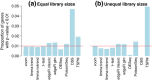
Figure 3
Type I error rates in the absence of true differential expression. The bar plots show the proportion of genes with P<0.01 for each method (a) when the library sizes are equal and (b) when the library sizes are unequal. The red line shows the nominal type I error rate of 0.01. Results are averaged over 100 simulations. Methods that control the type I error at or below the nominal level should lie below the red line.
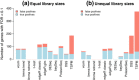
Figure 4
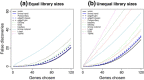
Power to detect true differential expression. Bars show the total number of genes that are detected as statistically significant (FDR < 0.1) (a) with equal library sizes and (b) with unequal library sizes. The blue segments show the number of true positives while the red segments show false positives. 200 genes are genuinely differentially expressed. Results are averaged over 100 simulations. Height of the blue bars shows empirical power. The ratio of the red to blue segments shows empirical FDR. FDR, false discovery rate.
Figure 5
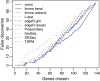
False discovery rates. The number of false discoveries is plotted for each method versus the number of genes selected as differentially expressed. Results are averaged over 100 simulations (a) with equal library sizes and (b) with unequal library sizes. voom has the lowest FDR at any cutoff in either scenario. FDR, false discovery rate.
Figure 6
False discovery rates evaluated from SEQC spike-in data. The number of false discoveries is plotted for each method versus the number of genes selected as differentially expressed. voom has the lowest false discovery rate overall.
Figure 7
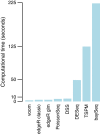
Computing times of RNA-seq methods. Bars show time in seconds required for the analysis of one simulated dataset on a MacBook laptop. Methods are ordered from quickest to most expensive.
Figure 8
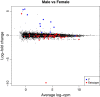
MA plot of male vs female comparison with male- and female-specific genes highlighted. The MA plot was produced by the limma plotMA function, and is a scatterplot of log-fold-change versus average log-cpm for each gene. Genes on the male-specific region of the Y chromosome genes are highlighted blue and are consistently upregulated in males, while genes on the X chromosome reported to escape X inactivation are highlighted red and are generally down in males. log-cpm, log-counts per million.
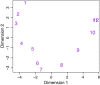
Figure 9
Multidimensional scaling plot of Drosophila melanogaster embryonic stages. Distances are computed from the log-cpm values. The 12 successive embryonic developmental stages are labeled 1 to 12, from earliest to latest.
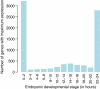
Figure 10
Number of genes associated with each Drosophila melanogaster embryonic stage. The number of genes whose peak estimated expression occurs at each of the stages is recorded.
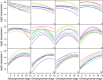
Figure 11
Expression trends for genes that peak at each Drosophila melanogaster embryonic stage. Panels (1) to (12) correspond to the 12 successive developmental stages. Each panel displays the fitted expression trends for the top ten genes that achieve their peak expression during that stage. In particular, panel (1) shows genes that are most highly expressed at the first stage and panel (12) shows genes most highly expressed at the last stage. Panels (7) and (8) are notable because they show genes with marked peaks at 12–14 hours and 14–16 hours respectively.
Similar articles
- Robust identification of differentially expressed genes from RNA-seq data.
Shahjaman M, Manir Hossain Mollah M, Rezanur Rahman M, Islam SMS, Nurul Haque Mollah M. Shahjaman M, et al. Genomics. 2020 Mar;112(2):2000-2010. doi: 10.1016/j.ygeno.2019.11.012. Epub 2019 Nov 20. Genomics. 2020. PMID: 31756426 - Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation.
McCarthy DJ, Chen Y, Smyth GK. McCarthy DJ, et al. Nucleic Acids Res. 2012 May;40(10):4288-97. doi: 10.1093/nar/gks042. Epub 2012 Jan 28. Nucleic Acids Res. 2012. PMID: 22287627 Free PMC article. - PLNseq: a multivariate Poisson lognormal distribution for high-throughput matched RNA-sequencing read count data.
Zhang H, Xu J, Jiang N, Hu X, Luo Z. Zhang H, et al. Stat Med. 2015 Apr 30;34(9):1577-89. doi: 10.1002/sim.6449. Epub 2015 Jan 30. Stat Med. 2015. PMID: 25641202 - Normalization for Single-Cell RNA-Seq Data Analysis.
Bacher R. Bacher R. Methods Mol Biol. 2019;1935:11-23. doi: 10.1007/978-1-4939-9057-3_2. Methods Mol Biol. 2019. PMID: 30758817 Review. - RNA-Seq methods for transcriptome analysis.
Hrdlickova R, Toloue M, Tian B. Hrdlickova R, et al. Wiley Interdiscip Rev RNA. 2017 Jan;8(1):10.1002/wrna.1364. doi: 10.1002/wrna.1364. Epub 2016 May 19. Wiley Interdiscip Rev RNA. 2017. PMID: 27198714 Free PMC article. Review.
Cited by
- Genetic and microenvironmental intra-tumor heterogeneity impacts colorectal cancer evolution and metastatic development.
Sobral D, Martins M, Kaplan S, Golkaram M, Salmans M, Khan N, Vijayaraghavan R, Casimiro S, Fernandes A, Borralho P, Ferreira C, Pinto R, Abreu C, Costa AL, Zhang S, Pawlowski T, Godsey J, Mansinho A, Macedo D, Lobo-Martins S, Filipe P, Esteves R, Coutinho J, Costa PM, Ramires A, Aldeia F, Quintela A, So A, Liu L, Grosso AR, Costa L. Sobral D, et al. Commun Biol. 2022 Sep 9;5(1):937. doi: 10.1038/s42003-022-03884-x. Commun Biol. 2022. PMID: 36085309 Free PMC article. - Schwann cell plasticity regulates neuroblastic tumor cell differentiation via epidermal growth factor-like protein 8.
Weiss T, Taschner-Mandl S, Janker L, Bileck A, Rifatbegovic F, Kromp F, Sorger H, Kauer MO, Frech C, Windhager R, Gerner C, Ambros PF, Ambros IM. Weiss T, et al. Nat Commun. 2021 Mar 12;12(1):1624. doi: 10.1038/s41467-021-21859-0. Nat Commun. 2021. PMID: 33712610 Free PMC article. - Tumor-associated neutrophils (TANs) in human carcinoma-draining lymph nodes: a novel TAN compartment.
Lonardi S, Missale F, Calza S, Bugatti M, Vescovi R, Debora B, Uppaluri R, Egloff AM, Mattavelli D, Lombardi D, Benerini Gatta L, Marini O, Tamassia N, Gardiman E, Cassatella MA, Scapini P, Nicolai P, Vermi W. Lonardi S, et al. Clin Transl Immunology. 2021 Feb 16;10(2):e1252. doi: 10.1002/cti2.1252. eCollection 2021. Clin Transl Immunology. 2021. PMID: 33643653 Free PMC article. - Sphingolipids, Transcription Factors, and Conserved Toolkit Genes: Developmental Plasticity in the Ant Cardiocondyla obscurior.
Schrader L, Simola DF, Heinze J, Oettler J. Schrader L, et al. Mol Biol Evol. 2015 Jun;32(6):1474-86. doi: 10.1093/molbev/msv039. Epub 2015 Feb 27. Mol Biol Evol. 2015. PMID: 25725431 Free PMC article. - Foxp3 enhancers synergize to maximize regulatory T cell suppressive capacity.
Zong X, Hao X, Xu B, Crawford JC, Wright S, Li J, Zhang Y, Bai L, He M, Jiang M, Fan Y, Connelly JP, Pruett-Miller SM, Berns H, Janke L, Li C, Feng Y. Zong X, et al. J Exp Med. 2021 Aug 2;218(8):e20202415. doi: 10.1084/jem.20202415. Epub 2021 Jun 4. J Exp Med. 2021. PMID: 34086055 Free PMC article.
References
- Smyth G. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:Article 3. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources