Structural mechanism of voltage-dependent gating in an isolated voltage-sensing domain - PubMed (original) (raw)
doi: 10.1038/nsmb.2768. Epub 2014 Feb 2.
Sherry Wanderling 1, Marcin Paduch 1, David Medovoy 1, Abhishek Singharoy 2, Ryan McGreevy 2, Carlos A Villalba-Galea 3, Raymond E Hulse 1, Benoît Roux 4, Klaus Schulten 5, Anthony Kossiakoff 4, Eduardo Perozo 4
Affiliations
- PMID: 24487958
- PMCID: PMC4116111
- DOI: 10.1038/nsmb.2768
Structural mechanism of voltage-dependent gating in an isolated voltage-sensing domain
Qufei Li et al. Nat Struct Mol Biol. 2014 Mar.
Abstract
The transduction of transmembrane electric fields into protein motion has an essential role in the generation and propagation of cellular signals. Voltage-sensing domains (VSDs) carry out these functions through reorientations of positive charges in the S4 helix. Here, we determined crystal structures of the Ciona intestinalis VSD (Ci-VSD) in putatively active and resting conformations. S4 undergoes an ~5-Å displacement along its main axis, accompanied by an ~60° rotation. This movement is stabilized by an exchange in countercharge partners in helices S1 and S3 that generates an estimated net charge transfer of ~1 eo. Gating charges move relative to a ''hydrophobic gasket' that electrically divides intra- and extracellular compartments. EPR spectroscopy confirms the limited nature of S4 movement in a membrane environment. These results provide an explicit mechanism for voltage sensing and set the basis for electromechanical coupling in voltage-dependent enzymes and ion channels.
Figures
Figure 1
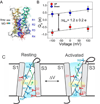
Stabilizing two conformations of Ci-VSD by local field engineering.A. Ci-VSP is composed of a voltage sensing domain and a phosphatase domain. Left, linear schematics of Ci-VSP sequence regions. Highlighted in red is the conserved S4 helix containing positive charges every three residues. The relative location the protein constructs for crystallization is shown on top: residues 89–260 (1) for R217E mutant and 106–244 (2) for WT. Right, general cartoon of Ci-VSD embedded in a lipid membrane. B. Representative traces of WT Ci-VSP gating (sensing) currents. The voltage dependence of Ci-VSP can be dramatically left shifted by mutations on the extracellular end of S4 at position 217: V½ = +58 mV for WT (black circle); − 15 mV for R217Q (white triangle); −62 mV for R217E (red triangle). Under 0 mV conditions, Ci-VSD populates the activated (“Up”) state for R217E mutant and the resting (“Down”) state for WT.C. Positive holding potential can promote transition of VSDs into “relaxed” state that generates a leftward shift of its voltage dependence. Left, basic kinetic cycle of canonical gating transitions. Right, voltage dependence of the transition into the relaxed state. Noticeable differences only take place for holding potentials higher than 20 mV. Therefore, at 0 mV, Ci-VSD WT and R217E mutants likely populate the canonical Down and Up states respectively.
Figure 2
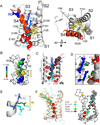
Structure of Ci-VSD R217E in the activated (“Up”) conformation.A. Side (left) and top (right) view of the Up conformation of Ci-VSD R217E in cartoon representation with S4 colored red. The four positive charges (R223, R226, R229 and R232) are depicted as sticks and colored in a series of blue shades. A well-defined hydrophobic gasket was formed by I126, F161, I190 with a gating charge R229 (R3).B. Architecture of the internal and external vestibules as calculated with the program HOLE61. The narrowest region is surrounded by the hydrophobic gasket at R3 (in yellow). C. With the subtle tilt and bend of S4, all four gating charges form a neat line perpendicular the hydrophobic gasket. The negative counter charges (D129:D1, D186:D2 and E183:E3) are located in S1 and S3. D. The top two arginines, R223 (R1) and R226 (R2) above the hydrophobic gasket are stabilized by hydrogen bonds with crystallographic waters between the negative charges D129 and D151.E. Left, heterogeneity of arginine positions within S4 was highlighted with VSD alignment in reference to Kv1.2 chimera (emerald): KvAP (yellow, RMSD=1.4 Å), NavRh (orange, RMSD=3.0 Å), NavAb (green, RMSD=1.1 Å) and Ci-VSD R217E (red, RMSD=1.1 Å). RMSD is calculated with Cα of aligned region (S1+S2+S3a) indicating the general agreement of VSD scaffold. The gating charges, shown in sphere, follow an arched path along S4 and but are distinctly different among the homologs even though all of them were expected to populate at Up state. Right, heterogeneity of putative counter charge positions within the VSD scaffold. Based on the crystal structures, only countercharges potentially involved in direct ion pairing with gating charges are shown.
Figure 3
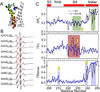
The nature of S4 rearrangement in a lipid environment. A. The region studied by site-directed spin labeling EPR spectroscopy covering the S3–S4 loop and entire S4 and the S4-phosphatase linker. B. Representative EPR spectra of the S4-phosphatase linker (residues 241–250) on both Ci-VSD WT (black) and R217E (red) background. C. Mobility, oxygen accessibility and NiEDDA accessibility of WT (black) and R217E (blue). The characteristic large ΠO2 oscillation and low ΠNi of transmembrane region are consistent with the boundary of S4 (shown on the top as reference) defined by crystal structure. Comparing the ΠNi, there is a decrease at the top of S4 with a simultaneous increase at the bottom of S4 from R217E to WT. This is consistent with a modest down movement of S4 from the Up state (R217E) to the Down state (WT). Residues 245 and 247 have significantly lower mobility and ΠNi in WT indicating stronger tertiary contact.
Figure 4
Structure of WT Ci-VSD in the resting (“Down”) conformation.A. Electron density map (2Fo-Fc at σt1.0) of WT Ci-VSD at 3.6 Å resolution. The backbone of S4 and inter-domain linker are colored red. B. Top: Three potential models within the electron density map of Ci-VSD WT: Two-click down (S4 in red), One-click down (S4 in green) and Up-conformation (S4 in blue). The rotation angle and vertical displacement of S4 were measured in reference to the One-click down model. The first Arginines R223 of three models were shown with yellow sphere and arrow to illustrate the vertical displacement on S4 helix. The independent R-free values and correlation coefficients were averaged among frames of every 2° rotation for simplicity.
Figure 5
Structural change between Ci-VSD WT and R217E. A. Overlap of Ci-VSD R217E (white) and WT (black) backbones reveals the conformational changes S3–S4 linker, relative position change in S4 and the existence of inter-domain linker in WT. The axial translations of Cα of the arginines are about ~5 Å. The four arginines rotates 48~75° relative to the helical axis. C. Ci-VSD WT has nearly identical scaffold (ribbon) and the conserved phenylalanine (sphere, highlighted in red circle), but a distinctly lower inter-domain linker than Kv1.2 chimera and NavAb. Based on the Z-axe established by potassium ions in Kv1.2 chimera filter, the Ci-VSD WT linker is ~ 2 Å lower on average and ~ 5 Å lower at the end of S4.
Figure 6
A molecular mechanism of charge translocation in voltage sensor. A. Comparison of the rotamer distribution of gating arginines in Up conformation resolved in the 2.5 Å crystal structure. There is a large downward reorientation going down the S4 and translocating through the hydrophobic gasket. B. Crystal structure of Ci-VSD WT (grey) is stable after 105 ns of all-atom MD simulation (spectrum).C. Calculation of the displacement charge between the Up and Down conformation of Ci-VSD at +100, 0 and −100 mV yielding a gating charge of 1.2 ± 0.2 eo, which is in agreement of the apparent valence z=1~1.5 estimated by the activation slope of gating current. D. Four existing metal bridges constrains for the down state in Shaker: bridge 1, 2 (Campos et al., 2007), bridge 3 (Lin et al., 2010) and bridge 4 (Tao et al., 2010). The three statedependent bridges 1, 2 and 4 are only compatible at Down state, but not the Up state of Ci-VSD. E. A mechanistic model of voltage sensing in Ci-VSD. A ~5 Å S4 down movement is accompanied by a counterclockwise ~60° rotation of the entire helix. arginines in S4 are stabilized by successive negative counter charges on S1 and S3. A potential rotameric reorientation of the arginines might additionally contribute to the overall gating charge transfer.
Similar articles
- Expression, purification, and reconstitution of the voltage-sensing domain from Ci-VSP.
Li Q, Jogini V, Wanderling S, Cortes DM, Perozo E. Li Q, et al. Biochemistry. 2012 Oct 16;51(41):8132-42. doi: 10.1021/bi300980q. Epub 2012 Oct 5. Biochemistry. 2012. PMID: 22989304 Free PMC article. - Probing α-3(10) transitions in a voltage-sensing S4 helix.
Kubota T, Lacroix JJ, Bezanilla F, Correa AM. Kubota T, et al. Biophys J. 2014 Sep 2;107(5):1117-1128. doi: 10.1016/j.bpj.2014.07.042. Biophys J. 2014. PMID: 25185547 Free PMC article. - Structural basis for gating charge movement in the voltage sensor of a sodium channel.
Yarov-Yarovoy V, DeCaen PG, Westenbroek RE, Pan CY, Scheuer T, Baker D, Catterall WA. Yarov-Yarovoy V, et al. Proc Natl Acad Sci U S A. 2012 Jan 10;109(2):E93-102. doi: 10.1073/pnas.1118434109. Epub 2011 Dec 12. Proc Natl Acad Sci U S A. 2012. PMID: 22160714 Free PMC article. - Voltage Sensing in Membranes: From Macroscopic Currents to Molecular Motions.
Freites JA, Tobias DJ. Freites JA, et al. J Membr Biol. 2015 Jun;248(3):419-30. doi: 10.1007/s00232-015-9805-x. Epub 2015 May 14. J Membr Biol. 2015. PMID: 25972106 Free PMC article. Review. - The EAG Voltage-Dependent K+ Channel Subfamily: Similarities and Differences in Structural Organization and Gating.
Barros F, de la Peña P, Domínguez P, Sierra LM, Pardo LA. Barros F, et al. Front Pharmacol. 2020 Apr 15;11:411. doi: 10.3389/fphar.2020.00411. eCollection 2020. Front Pharmacol. 2020. PMID: 32351384 Free PMC article. Review.
Cited by
- Free-energy landscape of ion-channel voltage-sensor-domain activation.
Delemotte L, Kasimova MA, Klein ML, Tarek M, Carnevale V. Delemotte L, et al. Proc Natl Acad Sci U S A. 2015 Jan 6;112(1):124-9. doi: 10.1073/pnas.1416959112. Epub 2014 Dec 22. Proc Natl Acad Sci U S A. 2015. PMID: 25535341 Free PMC article. - Voltage-Gated Potassium Channels: A Structural Examination of Selectivity and Gating.
Kim DM, Nimigean CM. Kim DM, et al. Cold Spring Harb Perspect Biol. 2016 May 2;8(5):a029231. doi: 10.1101/cshperspect.a029231. Cold Spring Harb Perspect Biol. 2016. PMID: 27141052 Free PMC article. Review. - BK channels: multiple sensors, one activation gate.
Yang H, Zhang G, Cui J. Yang H, et al. Front Physiol. 2015 Feb 6;6:29. doi: 10.3389/fphys.2015.00029. eCollection 2015. Front Physiol. 2015. PMID: 25705194 Free PMC article. Review. - Voltage sensitive phosphatases: emerging kinship to protein tyrosine phosphatases from structure-function research.
Hobiger K, Friedrich T. Hobiger K, et al. Front Pharmacol. 2015 Jan 10;6:20. doi: 10.3389/fphar.2015.00020. eCollection 2015. Front Pharmacol. 2015. PMID: 25713537 Free PMC article. Review. - From Nanodiscs to Isotropic Bicelles: A Procedure for Solution Nuclear Magnetic Resonance Studies of Detergent-Sensitive Integral Membrane Proteins.
Laguerre A, Löhr F, Henrich E, Hoffmann B, Abdul-Manan N, Connolly PJ, Perozo E, Moore JM, Bernhard F, Dötsch V. Laguerre A, et al. Structure. 2016 Oct 4;24(10):1830-1841. doi: 10.1016/j.str.2016.07.017. Epub 2016 Sep 15. Structure. 2016. PMID: 27618661 Free PMC article.
References
- Ahern CA, Horn R. Focused electric field across the voltage sensor of potassium channels. Neuron. 2005;48:25–29. - PubMed
- Armstrong CM, Bezanilla F. Currents related to movement of the gating particles of the sodium channels. Nature. 1973;242:459–461. - PubMed
- Bezanilla F. How membrane proteins sense voltage. Nat Rev Mol Cell Biol. 2008;9:323–332. - PubMed
Publication types
MeSH terms
Grants and funding
- R01-GM57846/GM/NIGMS NIH HHS/United States
- U01 GM094588/GM/NIGMS NIH HHS/United States
- RC2GM093307/GM/NIGMS NIH HHS/United States
- U54 GM087519/GM/NIGMS NIH HHS/United States
- T32 EB009412/EB/NIBIB NIH HHS/United States
- P41 GM104601/GM/NIGMS NIH HHS/United States
- RC2 GM093307/GM/NIGMS NIH HHS/United States
- U54 GM074946/GM/NIGMS NIH HHS/United States
- R01 GM062342/GM/NIGMS NIH HHS/United States
- KL2 TR000057/TR/NCATS NIH HHS/United States
- R01 GM072688/GM/NIGMS NIH HHS/United States
- UL1 TR000058/TR/NCATS NIH HHS/United States
- 9P41-GM104601/GM/NIGMS NIH HHS/United States
- R01 GM057846/GM/NIGMS NIH HHS/United States
- R01-GM062342/GM/NIGMS NIH HHS/United States
- U54-GM087519/GM/NIGMS NIH HHS/United States
- R01 GM098243/GM/NIGMS NIH HHS/United States
- U54-GM74946/GM/NIGMS NIH HHS/United States
- 5R01-GM098243-02/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources