Comparative functional genomics of Lactobacillus spp. reveals possible mechanisms for specialization of vaginal lactobacilli to their environment - PubMed (original) (raw)
Comparative Study
. 2014 Apr;196(7):1458-70.
doi: 10.1128/JB.01439-13. Epub 2014 Jan 31.
Affiliations
- PMID: 24488312
- PMCID: PMC3993339
- DOI: 10.1128/JB.01439-13
Comparative Study
Comparative functional genomics of Lactobacillus spp. reveals possible mechanisms for specialization of vaginal lactobacilli to their environment
Helena Mendes-Soares et al. J Bacteriol. 2014 Apr.
Abstract
Lactobacilli are found in a wide variety of habitats. Four species, Lactobacillus crispatus, L. gasseri, L. iners, and L. jensenii, are common and abundant in the human vagina and absent from other habitats. These may be adapted to the vagina and possess characteristics enabling them to thrive in that environment. Furthermore, stable codominance of multiple Lactobacillus species in a single community is infrequently observed. Thus, it is possible that individual vaginal Lactobacillus species possess unique characteristics that confer to them host-specific competitive advantages. We performed comparative functional genomic analyses of representatives of 25 species of Lactobacillus, searching for habitat-specific traits in the genomes of the vaginal lactobacilli. We found that the genomes of the vaginal species were significantly smaller and had significantly lower GC content than those of the nonvaginal species. No protein families were found to be specific to the vaginal species analyzed, but some were either over- or underrepresented relative to nonvaginal species. We also found that within the vaginal species, each genome coded for species-specific protein families. Our results suggest that even though the vaginal species show no general signatures of adaptation to the vaginal environment, each species has specific and perhaps unique ways of interacting with its environment, be it the host or other microbes in the community. These findings will serve as a foundation for further exploring the role of lactobacilli in the ecological dynamics of vaginal microbial communities and their ultimate impact on host health.
Figures
FIG 1
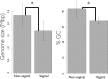
Genome size and GC content of Lactobacillus species. Data represent average genome size and % GC content of the Lactobacillus species isolated from the vagina and from other habitats (gastrointestinal tract and food). Error bars denote standard deviations (SD). The Kruskal-Wallis rank sum test was done on vaginal versus nonvaginal Lactobacillus strains to compare the two groups, and significant differences (P < 0.05) are denoted with an asterisk (*).
FIG 2
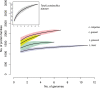
Gene number in Lactobacillus species. Gene accumulation rarefaction curves across Lactobacillus strains were calculated based on the presence or absence of protein families using the “specaccum” function in the “vegan” package of R (
) and were estimated by bootstrapping 100 permutations of randomized sample orders. The curves for all the genomes analyzed in this study (gray) and for the genomes of L. crispatus (pink), L. gasseri (yellow), L. jensenii (green), and L. iners (blue) are represented.
FIG 3
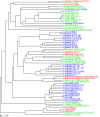
Hierarchical clustering of Lactobacillus strains based on dissimilarities in gene content. The dissimilarities were measured on binary data using Jaccard distance values, ranging from 0 to 1, to evaluate the presence or absence of each of the protein families. The bar on the left bottom of the figure represents a distance of 0.01. The different colors of the leaves represent the habitats from which the strains were isolated. Red represents strains isolated from food, green represents strains isolated from gastrointestinal tract, and blue represents strains isolated from the vagina.
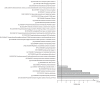
FIG 4
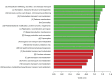
Overrepresentation (green) and underrepresentation (red) of the Clusters of Orthologous Genes (COGs) in the genomes of the vaginal Lactobacillus species. The horizontal scale at the bottom represents the odds ratio calculated for each COG.
FIG 5
Cluster of Orthologous Genes (COGs) with over- and underrepresentation in vaginal Lactobacillus species. Data represent Clusters of Orthologous Genes (COGs) that were significantly overrepresented (odds ratio > 1) and underrepresented (odds ratio < 1) in the vaginal relative to nonvaginal Lactobacillus species. CoA, coenzyme A.
Similar articles
- Genomic Comparisons of Lactobacillus crispatus and Lactobacillus iners Reveal Potential Ecological Drivers of Community Composition in the Vagina.
France MT, Mendes-Soares H, Forney LJ. France MT, et al. Appl Environ Microbiol. 2016 Nov 21;82(24):7063-7073. doi: 10.1128/AEM.02385-16. Print 2016 Dec 15. Appl Environ Microbiol. 2016. PMID: 27694231 Free PMC article. - Vaginal Microbiome of Pregnant Indian Women: Insights into the Genome of Dominant Lactobacillus Species.
Mehta O, Ghosh TS, Kothidar A, Gowtham MR, Mitra R, Kshetrapal P, Wadhwa N, Thiruvengadam R; GARBH-Ini study group; Nair GB, Bhatnagar S, Das B. Mehta O, et al. Microb Ecol. 2020 Aug;80(2):487-499. doi: 10.1007/s00248-020-01501-0. Epub 2020 Mar 23. Microb Ecol. 2020. PMID: 32206831 - Comparison of detection methods for vaginal lactobacilli.
Smidt I, Kiiker R, Oopkaup H, Lapp E, Rööp T, Truusalu K, Štšepetova J, Truu J, Mändar R. Smidt I, et al. Benef Microbes. 2015;6(5):747-51. doi: 10.3920/BM2014.0154. Epub 2015 Apr 22. Benef Microbes. 2015. PMID: 25869280 - Lactobacillus iners: Friend or Foe?
Petrova MI, Reid G, Vaneechoutte M, Lebeer S. Petrova MI, et al. Trends Microbiol. 2017 Mar;25(3):182-191. doi: 10.1016/j.tim.2016.11.007. Epub 2016 Dec 1. Trends Microbiol. 2017. PMID: 27914761 Review. - Vaginal microbiota and the potential of Lactobacillus derivatives in maintaining vaginal health.
Chee WJY, Chew SY, Than LTL. Chee WJY, et al. Microb Cell Fact. 2020 Nov 7;19(1):203. doi: 10.1186/s12934-020-01464-4. Microb Cell Fact. 2020. PMID: 33160356 Free PMC article. Review.
Cited by
- More Easily Cultivated Than Identified: Classical Isolation With Molecular Identification of Vaginal Bacteria.
Srinivasan S, Munch MM, Sizova MV, Fiedler TL, Kohler CM, Hoffman NG, Liu C, Agnew KJ, Marrazzo JM, Epstein SS, Fredricks DN. Srinivasan S, et al. J Infect Dis. 2016 Aug 15;214 Suppl 1(Suppl 1):S21-8. doi: 10.1093/infdis/jiw192. J Infect Dis. 2016. PMID: 27449870 Free PMC article. - Microbiota and Recurrent Pregnancy Loss (RPL); More than a Simple Connection.
Garmendia JV, De Sanctis CV, Hajdúch M, De Sanctis JB. Garmendia JV, et al. Microorganisms. 2024 Aug 10;12(8):1641. doi: 10.3390/microorganisms12081641. Microorganisms. 2024. PMID: 39203483 Free PMC article. Review. - Adverse effect of lactobacilli-depauperate cervicovaginal microbiota on pregnancy outcomes in women undergoing frozen-thawed embryo transfer.
Tsai HW, Tsui KH, Chiu YC, Wang LC. Tsai HW, et al. Reprod Med Biol. 2023 Jan 20;22(1):e12495. doi: 10.1002/rmb2.12495. eCollection 2023 Jan-Dec. Reprod Med Biol. 2023. PMID: 36699957 Free PMC article. - Comparative Genome Analysis of Bacillus amyloliquefaciens Focusing on Phylogenomics, Functional Traits, and Prevalence of Antimicrobial and Virulence Genes.
Liu H, Prajapati V, Prajapati S, Bais H, Lu J. Liu H, et al. Front Genet. 2021 Sep 30;12:724217. doi: 10.3389/fgene.2021.724217. eCollection 2021. Front Genet. 2021. PMID: 34659348 Free PMC article. - Alteration of vaginal microbiota in patients with unexplained recurrent miscarriage.
Zhang F, Zhang T, Ma Y, Huang Z, He Y, Pan H, Fang M, Ding H. Zhang F, et al. Exp Ther Med. 2019 May;17(5):3307-3316. doi: 10.3892/etm.2019.7337. Epub 2019 Mar 4. Exp Ther Med. 2019. PMID: 30988706 Free PMC article.
References
- Ravel J, Gajer P, Abdo Z, Schneider GM, Koenig SSK, McCulle SL, Karlebach S, Gorle R, Russel J, Tacket CO, Brotman RM, Davis CC, Ault K, Peralta L, Forney LJ. 2011. Vaginal microbiome of reproductive-age women. Proc. Natl. Acad. Sci. U. S. A. 108:4680–4687. 10.1073/pnas.1002611107 - DOI - PMC - PubMed
- De Backer E, Verhelst R, Verstraelen H, Alqumber MA, Burton JP, Tagg JR, Temmerman M, Vaneechoutte M. 2007. Quantitative determination by real-time PCR of four vaginal Lactobacillus species, Gardnerella vaginalis and Atopobium vaginae indicates an inverse relationship between L. gasseri and L. iners. BMC Microbiol. 7:115. 10.1186/1471-2180-7-115 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous