Genetic activation of ERK5 MAP kinase enhances adult neurogenesis and extends hippocampus-dependent long-term memory - PubMed (original) (raw)
Genetic activation of ERK5 MAP kinase enhances adult neurogenesis and extends hippocampus-dependent long-term memory
Wenbin Wang et al. J Neurosci. 2014.
Abstract
Recent studies have shown that inhibition of adult neurogenesis impairs the formation of hippocampus-dependent memory. However, it is not known whether increasing adult neurogenesis affects the persistence of hippocampus-dependent long-term memory. Furthermore, signaling mechanisms that regulate adult neurogenesis are not fully defined. We recently reported that the conditional and targeted knock-out of ERK5 MAP kinase in adult neurogenic regions of the mouse brain attenuates adult neurogenesis in the hippocampus and disrupts several forms of hippocampus-dependent memory. Here, we developed a gain-of-function knock-in mouse model to specifically activate endogenous ERK5 in the neurogenic regions of the adult brain. We report that the selective and targeted activation of ERK5 increases adult neurogenesis in the dentate gyrus by enhancing cell survival, neuronal differentiation, and dendritic complexity. Conditional ERK5 activation also improves the performance of challenging forms of spatial learning and memory and extends hippocampus-dependent long-term memory. We conclude that enhancing signal transduction of a single signaling pathway within adult neural stem/progenitor cells is sufficient to increase adult neurogenesis and improve the persistence of hippocampus-dependent memory. Furthermore, activation of ERK5 may provide a novel therapeutic target to improve long-term memory.
Keywords: ERK5; MAP kinase; adult neurogenesis; memory persistence; spatial learning.
Figures
Figure 1.
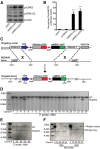
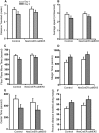
Strategy for generating (ROSA26)caMEK5-eGFPloxP knock-in mouse line. A, Biochemical analysis to confirm that the caMEK5-eGFP fusion protein is constitutively active and functional to activate ERK5. After transfection into NIH3T3 cells, caMEK5-eGFP fusion protein stimulates phosphorylation of endogenous ERK5, but not extracellular signal-regulated kinases 1/2 (ERK1/2). Empty vector (eGFP) was used as a negative control and caMEK5-HA as a positive control. β-actin was used as a loading control. B, caMEK5-eGFP fusion protein stimulates the transcriptional activity of MEF2C, a transcription factor and well characterized substrate of ERK5. NIH3T3 cells were transfected with a Gal4-luciferase reporter, an expression vector for Gal4-MEF2C fusion protein, and an expression vector for wild-type ERK5 to measure ERK5 stimulation of MEF2C-mediated transcriptional activity, as described previously (Cavanaugh et al., 2001). The activities of cells cotransfected with eGFP or wild-type (wt) MEK5 were used as baseline controls, whereas the activities of cells transfected with caMEK5-HA were used as a positive control. ***p < 0.001 compared with eGFP- or wt-MEK5-transfected cells. C, Schematic diagram depicting the caMEK5-eGFP targeting vector that targets the ROSA26 genomic locus after homologous recombination in mouse ES cells. cDNA sequence encoding caMEK5-eGFP fusion protein was cloned into the targeting vector behind a PGK-Neo-STOP cassette, which was flanked by two loxP sites. The targeting vector contains a CBA promoter, diphtheria toxin (PGK-DTA) for negative selection, and genomic ROSA26 sequences for homologous recombination. D, Southern blot analysis of DNA from 20 ES cell clones digested by MfeI and hybridized with a 3′ probe identified 2 targeted ES clones; clone #5 and clone #25. Of the 73 total ES cell clones screened, five were positive, targeted clones (data not shown). E, Southern analysis of DNA digested by NdeI and hybridized with a 5′ probe confirmed the five targeted clones. F, Southern blot analysis of DNA digested by MfeI or NdeI and hybridized with the Neo probe showed that three of the five ES cell clones were integrated with a single copy of the target gene: clones #25, #36, and #55. The ROSA caMEK5-eGFP mice were derived from clone #55.
Figure 2.
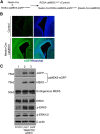
caMEK5-eGFP specifically activates endogenous ERK5, but not the closely related ERK1/2, in the brain after Cre-mediated recombination. A, Breeding scheme for generating Nestin-Cre:caMEK5 mice to express caMEK5-eGFP in the brain. (ROSA26)caMEK5-eGFPloxP/loxP mice were crossed with the Nestin-Cre mice to generate Nestin-Cre:caMEK5-eGFPloxP/+ mice, designated Nestin-Cre:caMEK5 mice. caMEK5-eGFPloxP/+ littermates were controls. B, Fluorescent images showing caMEK5-eGFP expression around the lateral ventricle regions of the brain of newborn Nestin-Cre:caMEK5 pups, but not that of the controls. Green, eGFP; blue, Hoechst. Scale bars, 100 μm in all images. C, Western blot analysis of newborn mouse brain lysates demonstrates expression of caMEK5-eGFP fusion protein in brain homogenates of Nestin-Cre:caMEK5 (pups #1 and #2), but not that of control littermate pups. The anti-MEK5 antibody recognizes both endogenous MEK5 and caMEK5-eGFP fusion proteins. Phosphorylated ERK5 (p-ERK5) and ERK1/2 (p-ERK1/2) were detected by an antibody that recognizes both on the same blot. Total ERK5 and β-actin were used as loading controls. Expression of caMEK5-eGFP fusion protein induced phosphorylation of endogenous ERK5, but not the closely related ERK1/2.
Figure 3.
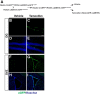
caMEK5-eGFP expression is conditionally induced in the adult neurogenic regions of the mouse brain. A, Experimental scheme for generating NesCreER:caMEK5 mice. Nestin-CreERTM:caMEK5-eGFPloxP/loxP mice were generated by crossing (ROSA26)caMEK5-eGFPloxP/loxP with Nestin-CreERTM mice. Eight-week-old mice were treated with vehicle (corn oil) or tamoxifen orally once a day for 7 d and perfused 3 weeks after the last tamoxifen dosing. B–I, Anti-eGFP immunostaining (green) demonstrates expression of caMEK5-eGFP in the SGZ of the DG and the SVZ of NesCreER:caMEK5, but not control mice. Scale bars, 50 μm in all panels.
Figure 4.
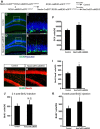
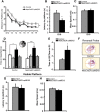
Induced conditional activation of ERK5 in the adult neurogenic regions of NesCreER:caMEK5 mice enhances hippocampal neurogenesis. A, Breeding and tamoxifen treatment scheme for generating NesCreER:caMEK5 and control littermates used in this and all behavior experiments. B–E, Representative DCX immunostaining images (green) from the SGZ of control (B, C) and NesCreER:caMEK5 (D, E) mouse brains, perfused 2 weeks after the last day of a 7 d tamoxifen treatment. Images in C and E are enlarged areas from boxed regions in B and D, respectively. Scale bars, 50 μm. F, Quantification of DCX+ cells in each DG. n = 4 mice/group; *p < 0.05. G, H, Representative BrdU (green) and NeuN (red) costaining from the SGZ of control (G) and NesCreER:caMEK5 (H) mouse brains perfused 7.5 weeks after the last day of a 7 d tamoxifen treatment. BrdU was injected once a day for 6 d starting 4 d after the last day of tamoxifen. Scale bar, 100 μm. I, Quantification of data from G and H, the number of NeuN+ and BrdU+ cells per DG. n = 5 mice/group; *p < 0.05. J, Ten days after the last tamoxifen injection, BrdU was injected every 2 h × 5 and mice were perfused 2 h after the last injection. Total BrdU+ cells in each DG of mice was quantified. n = 3–4 mice/group. K, Ten days after the last tamoxifen injection, BrdU was injected every 2 h × 5 and mice were perfused 6 weeks after the last BrdU injection. Total BrdU+ cells in each DG of mice were quantified. n = 5 mice/group; *p < 0.05. BrdU was injected every 2 h × 5 for J and K.
Figure 5.
Induced conditional caMEK5 expression in the adult neurogenic regions does not alter ERK1/2 activity. A, Representative phospho-ERK1/2 (p-ERK1/2) staining (green) in the DG and quantification of p-ERK1/2+ cells per DG. B, Representative phospho-ERK1/2 (p-ERK1/2) staining (green) in the pyramidal cell layer of the CA1 region and quantification of p-ERK1/2+ cells per CA1. n = 3 mice/group. N.S., not significant. Scale bar, 25 μm.
Figure 6.
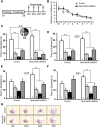
Expression of a single copy of the caMEK5-eGFP gene in the adult neurogenic regions of NesCreER:caMEK5/TdT mice is sufficient to increase hippocampal neurogenesis. A, Breeding and tamoxifen treatment scheme for generating NesCreER:caMEK5/TdT and control littermate mice. Mice were treated with tamoxifen daily for 7 d and perfused either 3 or 7 weeks later. B, C, Representative NeuN (green) and TdTomato (red) costaining from the SGZ of control (B) and NesCreER:caMEK5/TdT (C) mouse brains perfused 3 weeks after the last dose of tamoxifen. D, Quantification of NeuN+ and TdTomato+ (TdT+) cells per DG from data in B and C. E, F, Representative NeuN (green) and TdTomato (red) costaining from the SGZ of control (E) and NesCreER:caMEK5/TdT (F) mouse brains perfused 7 weeks after the last dose of tamoxifen. G, Quantification of NeuN+ and TdTomato+ (TdT+) cells per DG from data in E and F. H, I, Representative BrdU (green) and TdTomato (red) costaining from the SGZ of control (H) and NesCreER:caMEK5/TdT (I) mouse brains perfused 7 weeks after the last dose of tamoxifen. J, Quantification of data in H and I for the relative distribution of BrdU+ cells among recombined TdT+ cells versus BrdU+ cells among non-recombined TdT− cells. Scale bars, 100 μm. n = 4–5 mice/group; *p < 0.05; **p < 0.01.
Figure 7.
Induced conditional ERK5 activation enhances dendritic complexity of the adult-born neurons in the DG. Mice were treated with tamoxifen daily for 7 d and perfused 7 weeks later. A, Representative examples of Simple Neurite Tracer-generated traces of TdT+ neurons in control and NesCreER:caMEK5/TdT mice. B, Quantification of number of dendrites in TdT+ neurons. C, Average dendritic length of TdT+ neurons. D, Sholl analysis of TdT+ neurons. E, Representative confocal images showing spines of TdT+ neurons. F, Quantification of spine density of TdT+ neurons. Scale bars, 10 μm. n = 4–5 mice/group; N.S., not significant; *p < 0.05; **p < 0.01; ***p < 0.001.
Figure 8.
Inducible and conditional caMEK5 activation of ERK5 does not affect adult neural stem cell pool or the volume of the granular cell layer of the DG. A, B, Representative GFAP (green) and TdT (red) colabeling from the SGZ of control (A) and NesCreER:caMEK5/TdT (B) mouse brains perfused 7 weeks after the last dose of a 7 d tamoxifen treatment. Scale bar, 25 μm. C, Quantification of type I radial glia-like aNSC (GFAP+, TdT+ with radial glia-like morphology) in NesCreER:caMEK5/TdT and control mice. n = 4–5 mice/group. D, E, Representative NeuN (green) staining from the SGZ of control (D) and NesCreER:caMEK5/TdT (E) mouse brains perfused 7 weeks after the last dose of a 7 d tamoxifen treatment. Scale bar, 100 μm. F, Measurement of the granule cell layer volume in NesCreER:caMEK5/TdT and control mice using ImageJ (Sahay et al., 2011). n = 4–5 mice/group. N.S., not significant.
Figure 9.
NesCreER:caMEK5 and control mice have comparable basal locomotor activity in the open field tests. NesCreER:caMEK5 and control mice were treated with 3 cycles of a 4 d tamoxifen regimen with 2 weeks of intercycle interval. Behavior experiments were commenced 2 weeks after the last dose of tamoxifen. Mice were placed in an open field arena for 20 min each day for 2 d. A, Total distance traveled on d1 and d2. B, Average traveling speed. C, Floor plane move time. D, Total time spent along the margins of the arena. E, Total time spent in the center of the area. F, Percent distance traveled along the margin. n = 9–10/group. There was no statistical significant difference between the two groups of mice.
Figure 10.
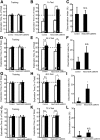
Conditional caMEK5 expression in adult neurogenic regions enhances spatial learning memory formation. A, Schematic depiction of the experimental design. Mice were subjected to the hidden platform water maze training (2 trials/d for 10 consecutive days, d1–d10), followed by a probe test 1 d later (d11). Mice were then subjected to reversal training (2 trials/d for 8 d, on d12–19) in which the hidden platform was relocated from Q1 to Q3. The reversal probe test was conducted 1 d after reversal training on d20 and the visible platform test on d22. B, During the 10 d training paradigm, NesCreER:caMEK5 mice swam significantly shorter distances than control mice to locate the hidden platform in Q1 on d9 and d10, suggesting enhanced spatial learning. C, Latency to platform on the last day of training (d10). D, The average swim speed quantified on d10. E, In the probe test, although both control and NesCreER:caMEK5 mice spent significantly more time in the virtual target quadrant, Q1, than any other three quadrants, NesCreER:caMEK5 mice spent even more time in Q1 than did control mice. One-way ANOVA analysis for control: F(3,36) = 32.7, p < 0.001; _post hoc_ analysis, training quadrant (Q1) > all other quadrants, p < 0.001. One-way ANOVA analysis for NesCreER:caMEK5: _F_(3,30) = 48.7, _p_ < 0.001; _post hoc_ analysis, training quadrant (Q1) > all other quadrants, p < 0.001. F, Representative ANYmaze program generated-swimming tracing pattern of control and NesCreER:caMEK5 mice in the probe tests. n = 9–10 mice/group. *p < 0.05; **p < 0.01; ***p < 0.001.
Figure 11.
Conditional caMEK5 expression in adult neurogenic regions improves reversal spatial learning memory formation. A, Mice were subjected to the reversal hidden platform water maze training after the probe test on d11, as depicted in Fig. 8_A_. During the 8 d reversal training (d12–d19), NesCreER:caMEK5 mice swam shorter distances than control mice to locate the hidden platform in Q3, suggesting enhanced reversal spatial learning. B, Analysis of the latency to platform on the last day of reversal training (d19) showed NesCreER:caMEK5 mice spent less time than control mice to find the hidden platform. C, The average swim speed on d19 training was comparable between the two groups of mice. D, In the reversal probe test, both control and NesCreER:caMEK5 mice spent significantly more time in the new virtual target quadrant Q3 than in the old target quadrant (Q1). E, The ratio of time spent in Q3 versus Q1 in reversal probe test was significantly higher for NesCreER:caMEK5 mice than control mice. F, Representative ANYmaze program generated-swimming tracing pattern of control and NesCreER:caMEK5 mice in the reversal probe tests. G, H, Control and NesCreER:caMEK5 mice performed equally well in the visible platform test; they showed comparable latency to acquisition of the platform location (G) and similar mean swimming speed (H). n = 9–10 mice/group. *p < 0.05; **p < 0.01; ***p < 0.001.
Figure 12.
Conditional caMEK5 expression in adult neurogenic regions extends long-term spatial memory. A, Schematic depiction of the experimental design. Mice were subjected to training for 8 d (d1–d8) with 4 trials/d in the hidden platform water maze. The first probe test was conducted on d9, the day after the last day of training, and second, third, and fourth probe tests were conducted every 2 weeks on d23, d37, and d51, respectively. B, During the 8 d training paradigm, target acquisition of the platform located in Q1 was similar for control and NesCreER:caMEK5 mice. C, Both control and NesCreER:caMEK5 mice spent significantly more time in Q1 than any other quadrants in the first probe test on d9. One-way ANOVA analysis for control: F(3,32) = 34.0, p < 0.001; post hoc analysis, time in training quadrant (Q1) was more than that of any other quadrants, p < 0.001. One-way ANOVA analysis for NesCreER:caMEK5 mice: F(3,28) = 62.5, p < 0.001; post hoc analysis, time in training quadrant (Q1) was more than that of any other quadrants, p < 0.001. There is no statistically significant difference in time spent in Q1 between the two groups of mice (p = 0.12), suggesting that the two groups learned equally well. D, In the second probe test conducted on d23, both control and NesCreER:caMEK5 mice still remembered Q1 and spent significantly more time in Q1 than any other quadrants. One-way ANOVA analysis for control: F(3,32) = 28.2, p < 0.001; post hoc analysis, time in training quadrant (Q1) was more than that of any other quadrants, p < 0.001. One-way ANOVA analysis for NesCreER:caMEK5 mice: F(3,28) = 30.3, p < 0.001; post hoc analysis, time in training quadrant (Q1) was more than that of any other quadrants, p < 0.001. E, During the third probe test on d37, NesCreER:caMEK5, but not control mice, spent significantly more time in Q1 than any other quadrants. One-way ANOVA analysis for NesCreER:caMEK5: F(3,28) = 31.3, p < 0.001; post hoc analysis, time in training quadrant (Q1) was more than that of any other quadrants, p < 0.001. One-way ANOVA analysis for control mice: F(3,32) = 5.4, p = 0.004; post hoc analysis, time in Q1 was only more than that of the opposite quadrant (Q3), p < 0.01. F, NesCreER:caMEK5 mice, but not control mice, retained spatial memory for Q1 even at 6 weeks after training when the fourth probe test was conducted on d51. One-way ANOVA analysis for NesCreER:caMEK5 mice: F(3,28) = 11.2, p < 0.001; post hoc analysis, time in Q1 was more than that of any other quadrants, p < 0.01. One-way ANOVA analysis for control mice: F(3,32) = 7.1, p = 0.001; post hoc analysis, time in Q1 was only more than that of Q3, p < 0.05. G, Representative swimming pattern traces in the four probe tests generated by the ANYmaze program. n = 8–9 mice/group. *p < 0.05; **p < 0.01; ***p < 0.001.
Figure 13.
NesCreER:caMEK5 mice have longer memory in the novel object recognition test. Mice were trained with 2 objects for 5 min and the percentage of time spent on exploring each object was quantified. Mice were then introduced to 2 objects at 1, 24, 48 or 72 h later, one familiar (object a, d, g, or j) and one novel (object c, f, i, or l) as labeled in the figure, and the percentage of time spent on each object during a 5 min test period and the discrimination index was quantified. A–C, Training and 1 h test. D–F, Training and 24 h test. G–I, Training and 48 h test. J–L, Training and 72 h test. n = 9–10/group. N.S., not significant; *p < 0.05; **p < 0.01; ***p < 0.001.
Figure 14.
NesCreER:caMEK5 mice have longer memory in the object location recognition test. A, Tamoxifen treatment scheme for generating NesCreER:caMEK5 and two groups of control mice from littermates. B, Mice were trained with two identical objects located at difference places (location a and b) for 5 min and the percentage of time spent on exploring each object was quantified. At 1, 48, or 72 h after training, mice were introduced to the same two objects where one remained at the same location (location a) and the other moved to a novel location (location c). The percentage of time spent on each object during a 5 min test period and the discrimination index were quantified. B–D, Training and 1 h test. E–G, Training and 48 h test. H–J, Training and 72 h test. n = 10–11/group. N.S., not significant; *p < 0.05; **p < 0.01; ***p < 0.001.
Similar articles
- Inducible and conditional deletion of extracellular signal-regulated kinase 5 disrupts adult hippocampal neurogenesis.
Pan YW, Zou J, Wang W, Sakagami H, Garelick MG, Abel G, Kuo CT, Storm DR, Xia Z. Pan YW, et al. J Biol Chem. 2012 Jul 6;287(28):23306-17. doi: 10.1074/jbc.M112.344762. Epub 2012 May 29. J Biol Chem. 2012. PMID: 22645146 Free PMC article. - Inducible activation of ERK5 MAP kinase enhances adult neurogenesis in the olfactory bulb and improves olfactory function.
Wang W, Lu S, Li T, Pan YW, Zou J, Abel GM, Xu L, Storm DR, Xia Z. Wang W, et al. J Neurosci. 2015 May 20;35(20):7833-49. doi: 10.1523/JNEUROSCI.3745-14.2015. J Neurosci. 2015. PMID: 25995470 Free PMC article. - Role of adult neurogenesis in hippocampus-dependent memory, contextual fear extinction and remote contextual memory: new insights from ERK5 MAP kinase.
Pan YW, Storm DR, Xia Z. Pan YW, et al. Neurobiol Learn Mem. 2013 Oct;105:81-92. doi: 10.1016/j.nlm.2013.07.011. Epub 2013 Jul 18. Neurobiol Learn Mem. 2013. PMID: 23871742 Free PMC article. - The maintenance of established remote contextual fear memory requires ERK5 MAP kinase and ongoing adult neurogenesis in the hippocampus.
Pan YW, Storm DR, Xia Z. Pan YW, et al. PLoS One. 2012;7(11):e50455. doi: 10.1371/journal.pone.0050455. Epub 2012 Nov 26. PLoS One. 2012. PMID: 23189204 Free PMC article.
Cited by
- Can exercise benefits be harnessed with drugs? A new way to combat neurodegenerative diseases by boosting neurogenesis.
Zhao R. Zhao R. Transl Neurodegener. 2024 Jul 25;13(1):36. doi: 10.1186/s40035-024-00428-7. Transl Neurodegener. 2024. PMID: 39049102 Free PMC article. Review. - Conditional deletion of Ndufs4 in dopaminergic neurons promotes Parkinson's disease-like non-motor symptoms without loss of dopamine neurons.
Choi WS, Kim HW, Tronche F, Palmiter RD, Storm DR, Xia Z. Choi WS, et al. Sci Rep. 2017 Mar 22;7:44989. doi: 10.1038/srep44989. Sci Rep. 2017. PMID: 28327638 Free PMC article. - Nodakenin Enhances Cognitive Function and Adult Hippocampal Neurogenesis in Mice.
Gao Q, Jeon SJ, Jung HA, Lee HE, Park SJ, Lee Y, Lee Y, Ko SY, Kim B, Choi JS, Ryu JH. Gao Q, et al. Neurochem Res. 2015 Jul;40(7):1438-47. doi: 10.1007/s11064-015-1612-3. Epub 2015 May 22. Neurochem Res. 2015. PMID: 25998887 - The Perfect Cytokine Storm: How Peripheral Immune Challenges Impact Brain Plasticity & Memory Function in Aging.
Muscat SM, Barrientos RM. Muscat SM, et al. Brain Plast. 2021 Aug 23;7(1):47-60. doi: 10.3233/BPL-210127. eCollection 2021. Brain Plast. 2021. PMID: 34631420 Free PMC article. Review. - The effect of chronic stimulation of serotonin receptor type 7 on recognition, passive avoidance memory, hippocampal long-term potentiation, and neuronal apoptosis in the amyloid β protein treated rat.
Shahidi S, Asl SS, Komaki A, Hashemi-Firouzi N. Shahidi S, et al. Psychopharmacology (Berl). 2018 May;235(5):1513-1525. doi: 10.1007/s00213-018-4862-3. Epub 2018 Apr 10. Psychopharmacology (Berl). 2018. PMID: 29637287
References
- Birch AM, McGarry NB, Kelly AM. Short-term environmental enrichment, in the absence of exercise, improves memory, and increases NGF concentration, early neuronal survival, and synaptogenesis in the dentate gyrus in a time-dependent manner. Hippocampus. 2013;23:437–450. doi: 10.1002/hipo.22103. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 NS20498/NS/NINDS NIH HHS/United States
- P30 HD002274/HD/NICHD NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- R01 MH095840/MH/NIMH NIH HHS/United States
- U54 HD083091/HD/NICHD NIH HHS/United States
- R01 NS020498/NS/NINDS NIH HHS/United States
- P30 HD02274/HD/NICHD NIH HHS/United States
- R01 MH95840/MH/NIMH NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous