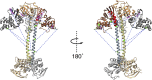Single-particle EM reveals the higher-order domain architecture of soluble guanylate cyclase - PubMed (original) (raw)
Single-particle EM reveals the higher-order domain architecture of soluble guanylate cyclase
Melody G Campbell et al. Proc Natl Acad Sci U S A. 2014.
Abstract
Soluble guanylate cyclase (sGC) is the primary nitric oxide (NO) receptor in mammals and a central component of the NO-signaling pathway. The NO-signaling pathways mediate diverse physiological processes, including vasodilation, neurotransmission, and myocardial functions. sGC is a heterodimer assembled from two homologous subunits, each comprised of four domains. Although crystal structures of isolated domains have been reported, no structure is available for full-length sGC. We used single-particle electron microscopy to obtain the structure of the complete sGC heterodimer and determine its higher-order domain architecture. Overall, the protein is formed of two rigid modules: the catalytic dimer and the clustered Per/Art/Sim and heme-NO/O2-binding domains, connected by a parallel coiled coil at two hinge points. The quaternary assembly demonstrates a very high degree of flexibility. We captured hundreds of individual conformational snapshots of free sGC, NO-bound sGC, and guanosine-5'-[(α,β)-methylene]triphosphate-bound sGC. The molecular architecture and pronounced flexibility observed provides a significant step forward in understanding the mechanism of NO signaling.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
sGC domain organization and X-ray crystallographic models. Each subunit contains four modular domains; α1 domains are shown in shades of gray, and β1 domains are shown in color. The H-NOX domain of the β1 subunit contains the heme cofactor, shown in red. Structures for Rattus norvegicus are modeled based on previously solved crystal structures of homologous domains (Materials and Methods). The H-NOX structures are modeled from a standalone Nostoc sp. PCC 7120 H-NOX domain (PDB: 2O09) (6). The PAS and helical domains are modeled on individual domain truncations. The PAS domain is based on the PAS domain from Manduca sexta (PDB: 4GJ4) (7), and the helical domain is based on the β1 R. norvegicus structure (3HLS) (8). The catalytic domain is the Homo sapiens α1β1 crystal structure (PDB: 3UVJ) (9).
Fig. 2.
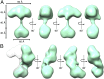
A continuous range of conformations is observed for the sGC holoenzyme, as illustrated here by 18 different conformational snapshots. Three-dimensional reconstructions are shown in green directly above the corresponding 2D class averages. The 3D maps were aligned relative to each other based on the heart-shaped catalytic domain. For consistency, models are displayed such that the four-lobed density is flexing toward the right, although we suggest that the range of motions allows the four-lobed cluster to make contact with both sides of the heart-shaped catalytic domain. (A) Nine snapshots of sGC exhibiting in-plane flexing of the four-lobed cluster. (B) Nine conformational snapshots exhibiting out-of-plane flexing of the four-lobed cluster. The thin stalk connecting the main densities is readily observed in the 2D class averages but sometimes only weakly observed in the 3D density maps, which are reconstructed from images tilted to a high angle (Materials and Methods).
Fig. 3.
Two selected sGC maps illustrating two extreme sGC conformations. (A) An extended structure showing the heart-shaped domain and the four-lobed cluster separated by 40 Å. (B) A “bent” structure where these two densities directly interact.
Fig. 4.
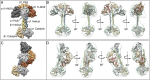
The higher-order domain architecture of sGC. Homology models of the individual domains were fit into representative EM maps exhibiting two conformational extremes: the extended conformation (A and B) and a bent conformation (C and D). The catalytic dimer fits in the heart-shaped density, and a parallel coiled coil connects it to the PAS/H-NOX cluster. The heme-associated helix (red) of the β1 H-NOX domain is positioned to interact with PAS dimer. The β1 H-NOX domain is also poised to interact directly with the α1 catalytic domain in the bent conformation. (A and C) Space-filling representation of the complete sGC structure emphasizing the striking consistency with the EM density. (B and D) Fit of the homology models (displayed as ribbons) within the EM density map.
Fig. 5.
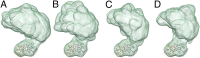
Merged maps illustrating the range of motion available to sGC when free or ligand-bound. Maps were aligned to the catalytic domain and show a similar range of motion under several experimental conditions. (A) The sGC holoenzyme. (B) NO-bound sGC. (C) GPCPP-bound sGC. (D) GPCPP and NO-bound sGC.
Fig. 6.
Previously found cross-links mapped onto model. Domains colored as previously described. Cross-links are shown as blue dashed lines. Residues contributing to cross-links within the PAS/H-NOX cluster are shown in magenta. Residues contributing to cross-links between the PAS/H-NOX cluster and the coiled coil are shown in green. The conformational space sampled by sGC in our reconstructions allows rationalizing a large amount of available cross-linking data when taking into account the fact that not all cross-links with the helical domains occur simultaneously.
Similar articles
- Higher-order interactions bridge the nitric oxide receptor and catalytic domains of soluble guanylate cyclase.
Underbakke ES, Iavarone AT, Marletta MA. Underbakke ES, et al. Proc Natl Acad Sci U S A. 2013 Apr 23;110(17):6777-82. doi: 10.1073/pnas.1301934110. Epub 2013 Apr 9. Proc Natl Acad Sci U S A. 2013. PMID: 23572573 Free PMC article. - Probing domain interactions in soluble guanylate cyclase.
Derbyshire ER, Winter MB, Ibrahim M, Deng S, Spiro TG, Marletta MA. Derbyshire ER, et al. Biochemistry. 2011 May 24;50(20):4281-90. doi: 10.1021/bi200341b. Epub 2011 May 3. Biochemistry. 2011. PMID: 21491957 Free PMC article. - Crystal structure of the Alpha subunit PAS domain from soluble guanylyl cyclase.
Purohit R, Weichsel A, Montfort WR. Purohit R, et al. Protein Sci. 2013 Oct;22(10):1439-44. doi: 10.1002/pro.2331. Epub 2013 Sep 7. Protein Sci. 2013. PMID: 23934793 Free PMC article. - Structure and Activation of Soluble Guanylyl Cyclase, the Nitric Oxide Sensor.
Montfort WR, Wales JA, Weichsel A. Montfort WR, et al. Antioxid Redox Signal. 2017 Jan 20;26(3):107-121. doi: 10.1089/ars.2016.6693. Epub 2016 Apr 26. Antioxid Redox Signal. 2017. PMID: 26979942 Free PMC article. Review. - Structure and regulation of soluble guanylate cyclase.
Derbyshire ER, Marletta MA. Derbyshire ER, et al. Annu Rev Biochem. 2012;81:533-59. doi: 10.1146/annurev-biochem-050410-100030. Epub 2012 Feb 9. Annu Rev Biochem. 2012. PMID: 22404633 Review.
Cited by
- Allosteric activation of the nitric oxide receptor soluble guanylate cyclase mapped by cryo-electron microscopy.
Horst BG, Yokom AL, Rosenberg DJ, Morris KL, Hammel M, Hurley JH, Marletta MA. Horst BG, et al. Elife. 2019 Sep 30;8:e50634. doi: 10.7554/eLife.50634. Elife. 2019. PMID: 31566566 Free PMC article. - Structural Perspectives on the Mechanism of Soluble Guanylate Cyclase Activation.
Wittenborn EC, Marletta MA. Wittenborn EC, et al. Int J Mol Sci. 2021 May 21;22(11):5439. doi: 10.3390/ijms22115439. Int J Mol Sci. 2021. PMID: 34064029 Free PMC article. Review. - Cryo-EM density map fitting driven in-silico structure of human soluble guanylate cyclase (hsGC) reveals functional aspects of inter-domain cross talk upon NO binding.
Khalid RR, Maryam A, Fadouloglou VE, Siddiqi AR, Zhang Y. Khalid RR, et al. J Mol Graph Model. 2019 Jul;90:109-119. doi: 10.1016/j.jmgm.2019.04.009. Epub 2019 Apr 24. J Mol Graph Model. 2019. PMID: 31055154 Free PMC article. - Sources of Vascular Nitric Oxide and Reactive Oxygen Species and Their Regulation.
Tejero J, Shiva S, Gladwin MT. Tejero J, et al. Physiol Rev. 2019 Jan 1;99(1):311-379. doi: 10.1152/physrev.00036.2017. Physiol Rev. 2019. PMID: 30379623 Free PMC article. Review. - Instability in a coiled-coil signaling helix is conserved for signal transduction in soluble guanylyl cyclase.
Weichsel A, Kievenaar JA, Curry R, Croft JT, Montfort WR. Weichsel A, et al. Protein Sci. 2019 Oct;28(10):1830-1839. doi: 10.1002/pro.3707. Epub 2019 Aug 27. Protein Sci. 2019. PMID: 31411784 Free PMC article.
References
- Lucas KA, et al. Guanylyl cyclases and signaling by cyclic GMP. Pharmacol Rev. 2000;52(3):375–414. - PubMed
- Bredt DS. Endogenous nitric oxide synthesis: Biological functions and pathophysiology. Free Radic Res. 1999;31(6):577–596. - PubMed
- Conole D, Scott LJ. Riociguat: First global approval. Drugs. 2013;73(17):1967–1975. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- F32 GM093564/GM/NIGMS NIH HHS/United States
- P41 GM103310/GM/NIGMS NIH HHS/United States
- 5F32GM093564/GM/NIGMS NIH HHS/United States
- 9 P41 GM103310/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources