BayMeth: improved DNA methylation quantification for affinity capture sequencing data using a flexible Bayesian approach - PubMed (original) (raw)
BayMeth: improved DNA methylation quantification for affinity capture sequencing data using a flexible Bayesian approach
Andrea Riebler et al. Genome Biol. 2014.
Abstract
Affinity capture of DNA methylation combined with high-throughput sequencing strikes a good balance between the high cost of whole genome bisulfite sequencing and the low coverage of methylation arrays. We present BayMeth, an empirical Bayes approach that uses a fully methylated control sample to transform observed read counts into regional methylation levels. In our model, inefficient capture can readily be distinguished from low methylation levels. BayMeth improves on existing methods, allows explicit modeling of copy number variation, and offers computationally efficient analytical mean and variance estimators. BayMeth is available in the Repitools Bioconductor package.
Figures
Figure 1
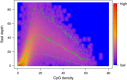
SssI read depth versus CpG density together with prior predictive distribution. Smoothed color density representation of SssI read depth versus CpG density together with the mean (green solid line) and 2.5_%_ and 97.5_%_ quantiles (green dashed lines) of the prior predictive distribution for the SssI control sample. The parameters for this negative binomial distribution were derived using an empirical Bayes approach by maximizing the joint marginal distribution of the IMR-90 and SssI control counts stratified into 100 CpG density groups. Only counts from bins with a mappability larger than 0.75 were considered.
Figure 2
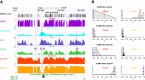
Example data tracks for IMR-90 chromosome 7. (A) WGBS methylome (black) per CpG-site and per 100-bp bin (purple) as obtained by Lister and others [40]. CpG density (light blue), and read counts for SssI-treated DNA (blue) and IMR-90 cells (green) obtained by MBD-seq based on 100-bp non-overlapping bins are shown. Methylation estimates for BayMeth (red) and Batman (orange) are provided. (B) Detailed posterior information for BayMeth and Batman for four specific bins of panel A (denoted a, b, c and d). For BayMeth, the posterior marginals together with 95% HPD credible intervals (shaded gray) are shown. The posterior samples obtained by Batman are plotted as histograms. For both approaches the posterior mean is indicated (red dashed line) together with the true WGBS-derived methylation estimate (blue dashed line). chr, chromosome; kb, kilobase; WGBS, whole genome bisulfite sequencing.
Figure 3
Regional methylation estimates for IMR-90 chromosome 7. Smoothed color density representation of regional DNAme estimates for BALM, MEDIPS, Batman, BayMeth and BayMeth ignoring SssI information, plotted against WGBS methylation levels for the 75% of bins with the largest depth in the truth (cutoff was 33 reads) where the depth in the SssI control was (27,168]. In addition the y = x line (green dashed line) is shown. Black points indicate outliers. WGBS, whole genome bisulfite sequencing.
Figure 4
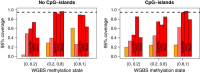
Coverage probabilities stratified by CpG island status and true methylation level. Coverage probabilities (frequency in which the true value is within a predefined credible interval) at the 95% level are shown for the 75% of bins with the largest depth in the truth (cutoff was 33 reads) for Batman (orange), BayMeth ignoring SssI control information (light red) and BayMeth (red). Three different types of credible intervals (quantile-based, Wald and HPD) are shown for BayMeth, while for Batman and the SssI-free version of BayMeth only quantile-based intervals are available. MEDIPS and BALM do not return any uncertainty estimates. The nominal coverage value is indicated (black dashed line) as a reference. Genomic regions were stratified by CpG density using the threshold of 12.46, which separates CpG islands from non-CpG islands; compare Additional file 2: Figure S1. Further stratification by the true methylation level as derived from WGBS [40] is provided. HPD, highest posterior density; WGBS, whole genome bisulfite sequencing.
Figure 5
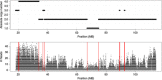
Relation between copy number state and regional affinity enrichment. Top: Copy number estimates for the LNCaP cell line obtained by the PICNIC [55] algorithm for 100-bp bins across human chromosome 13 with a mappability of at least 75%. Bottom: Read counts of affinity capture sequencing data for the same bins. MB, megabase.
Figure 6
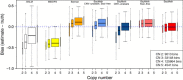
Bias of LNCaP methylation estimates compared to 450k array beta values. Box plots for bias (estimated methylation level minus 450K array beta value) for BALM (white), MEDIPS (yellow), Batman (orange), CNV-unaware and SssI-free BayMeth (light blue), CNV-unaware BayMeth (dark blue), SssI-free but CNV-aware BayMeth (light red) and CNV-aware BayMeth (red) stratified by copy numbers 2 to 5. (Outliers are not shown.) The width of the boxes is proportional to the percentage of bins (the legend gives the absolute numbers) for the copy number class. A uniform prior for the methylation level was used taking SssI information into account. In the SssI-free version a Dirac-Beta-Dirac mixture with weights fixed to 0.1, 0.8 and 0.1 was used. The results are shown genome-wide for 100-bp bins with at least 75% mappability and where the true methylation estimate is larger than 0.5. A threshold of 13 was applied for the depth of SssI. The blue dashed line indicates a bias of zero. CN, copy number; CNV, copy number variation.
Figure 7
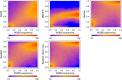
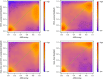
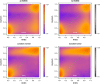
Effect of adjusting for CNV for the LNCaP cell line. Smoothed color density representation of methylation estimates for copy number state 2 derived by BayMeth compared to 450k array beta values. A threshold of 13 was applied for the depth of SssI, which gives 61,969 bins, of which we have for 18,010 100-bp bins a beta value and BayMeth estimate. In addition the y = x line (green dashed line) is shown. Black points indicate outliers. Top left: CNV-unaware BayMeth; top right: CNV-aware BayMeth; bottom left: SssI-free and CNV-unaware BayMeth; bottom right: SssI-free BayMeth. CNV, copy number variation.
Figure 8
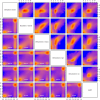
Comparison of raw IMR-90 data and methylation estimates obtained by different methylation kits. Genomic bins (100 bp) with a mappability larger than 75% for which the predicted HPD credible interval width was smaller than 0.4 were selected. For these bins the upper triangle of panels shows the smoothed color density (from blue for low density to red for high density) of the raw counts and the lower triangle of panels shows the estimated methylation levels obtained by different methylation kits against each other. The number of bins is given in white in the panels in the lower triangle.
Figure 9
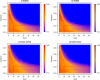
Regional methylation estimates for samples of Bock data. Smoothed color density representation of regional DNAme estimates of BayMeth, plotted against RRBS methylation levels, where the estimated standard deviation of BayMeth is smaller than 0.15 for bins with more than 20 reads for RRBS and at least a depth of 10 in the SssI control. (a)-(d) Single samples from the original study. The number of bins for each sample is shown at the bottom center of the panels. RRBS, reduced representation bisulfite sequencing.
Figure 10
Regional variance estimates versus SssI control for Bock data. Smoothed color density representation of variance estimates obtained by BayMeth versus number of reads in the SssI control for a read depth larger than 20 in RRBS. The red boxes contain the bins used in Figure 9, which have a depth of at least 10 in SssI and a standard deviation smaller than 0.15, i.e. a variance smaller than 0.025. (a)-(d) Single samples from the original study.
Similar articles
- A model of pulldown alignments from SssI-treated DNA improves DNA methylation prediction.
Moreland BS, Oman KM, Bundschuh R. Moreland BS, et al. BMC Bioinformatics. 2019 Aug 19;20(1):431. doi: 10.1186/s12859-019-3011-2. BMC Bioinformatics. 2019. PMID: 31426747 Free PMC article. - A full Bayesian partition model for identifying hypo- and hyper-methylated loci from single nucleotide resolution sequencing data.
Wang H, He C, Kushwaha G, Xu D, Qiu J. Wang H, et al. BMC Bioinformatics. 2016 Jan 11;17 Suppl 1(Suppl 1):7. doi: 10.1186/s12859-015-0850-3. BMC Bioinformatics. 2016. PMID: 26818685 Free PMC article. - A Flexible, Efficient Binomial Mixed Model for Identifying Differential DNA Methylation in Bisulfite Sequencing Data.
Lea AJ, Tung J, Zhou X. Lea AJ, et al. PLoS Genet. 2015 Nov 24;11(11):e1005650. doi: 10.1371/journal.pgen.1005650. eCollection 2015 Nov. PLoS Genet. 2015. PMID: 26599596 Free PMC article. - Methodological aspects of whole-genome bisulfite sequencing analysis.
Adusumalli S, Mohd Omar MF, Soong R, Benoukraf T. Adusumalli S, et al. Brief Bioinform. 2015 May;16(3):369-79. doi: 10.1093/bib/bbu016. Epub 2014 May 27. Brief Bioinform. 2015. PMID: 24867940 Review. - Analyzing the cancer methylome through targeted bisulfite sequencing.
Lee EJ, Luo J, Wilson JM, Shi H. Lee EJ, et al. Cancer Lett. 2013 Nov 1;340(2):171-8. doi: 10.1016/j.canlet.2012.10.040. Epub 2012 Nov 28. Cancer Lett. 2013. PMID: 23200671 Free PMC article. Review.
Cited by
- Statistical methods for detecting differentially methylated loci and regions.
Robinson MD, Kahraman A, Law CW, Lindsay H, Nowicka M, Weber LM, Zhou X. Robinson MD, et al. Front Genet. 2014 Sep 16;5:324. doi: 10.3389/fgene.2014.00324. eCollection 2014. Front Genet. 2014. PMID: 25278959 Free PMC article. Review. - Techniques and Approaches to Genetic Analyses in Nephrological Disorders.
Willig LK. Willig LK. J Pediatr Genet. 2016 Mar;5(1):2-14. doi: 10.1055/s-0035-1557108. Epub 2015 Aug 13. J Pediatr Genet. 2016. PMID: 27617137 Free PMC article. Review. - A probabilistic generative model for quantification of DNA modifications enables analysis of demethylation pathways.
Äijö T, Huang Y, Mannerström H, Chavez L, Tsagaratou A, Rao A, Lähdesmäki H. Äijö T, et al. Genome Biol. 2016 Mar 14;17:49. doi: 10.1186/s13059-016-0911-6. Genome Biol. 2016. PMID: 26975309 Free PMC article. - Benchmarking long-read RNA-sequencing analysis tools using in silico mixtures.
Dong X, Du MRM, Gouil Q, Tian L, Jabbari JS, Bowden R, Baldoni PL, Chen Y, Smyth GK, Amarasinghe SL, Law CW, Ritchie ME. Dong X, et al. Nat Methods. 2023 Nov;20(11):1810-1821. doi: 10.1038/s41592-023-02026-3. Epub 2023 Oct 2. Nat Methods. 2023. PMID: 37783886 - Genome-Wide Epigenetic Studies in Human Disease: A Primer on -Omic Technologies.
Yan H, Tian S, Slager SL, Sun Z, Ordog T. Yan H, et al. Am J Epidemiol. 2016 Jan 15;183(2):96-109. doi: 10.1093/aje/kwv187. Epub 2015 Dec 30. Am J Epidemiol. 2016. PMID: 26721890 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases