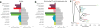The genome of a Late Pleistocene human from a Clovis burial site in western Montana - PubMed (original) (raw)
. 2014 Feb 13;506(7487):225-9.
doi: 10.1038/nature13025.
Sarah L Anzick 2, Michael R Waters 3, Pontus Skoglund 4, Michael DeGiorgio 5, Thomas W Stafford Jr 6, Simon Rasmussen 7, Ida Moltke 8, Anders Albrechtsen 9, Shane M Doyle 10, G David Poznik 11, Valborg Gudmundsdottir 7, Rachita Yadav 7, Anna-Sapfo Malaspinas 12, Samuel Stockton White 5th 13, Morten E Allentoft 12, Omar E Cornejo 14, Kristiina Tambets 15, Anders Eriksson 16, Peter D Heintzman 17, Monika Karmin 15, Thorfinn Sand Korneliussen 12, David J Meltzer 18, Tracey L Pierre 12, Jesper Stenderup 12, Lauri Saag 15, Vera M Warmuth 19, Margarida C Lopes 20, Ripan S Malhi 21, Søren Brunak 7, Thomas Sicheritz-Ponten 7, Ian Barnes 22, Matthew Collins 23, Ludovic Orlando 12, Francois Balloux 24, Andrea Manica 25, Ramneek Gupta 7, Mait Metspalu 15, Carlos D Bustamante 26, Mattias Jakobsson 27, Rasmus Nielsen 28, Eske Willerslev 12
Affiliations
- PMID: 24522598
- PMCID: PMC4878442
- DOI: 10.1038/nature13025
The genome of a Late Pleistocene human from a Clovis burial site in western Montana
Morten Rasmussen et al. Nature. 2014.
Abstract
Clovis, with its distinctive biface, blade and osseous technologies, is the oldest widespread archaeological complex defined in North America, dating from 11,100 to 10,700 (14)C years before present (bp) (13,000 to 12,600 calendar years bp). Nearly 50 years of archaeological research point to the Clovis complex as having developed south of the North American ice sheets from an ancestral technology. However, both the origins and the genetic legacy of the people who manufactured Clovis tools remain under debate. It is generally believed that these people ultimately derived from Asia and were directly related to contemporary Native Americans. An alternative, Solutrean, hypothesis posits that the Clovis predecessors emigrated from southwestern Europe during the Last Glacial Maximum. Here we report the genome sequence of a male infant (Anzick-1) recovered from the Anzick burial site in western Montana. The human bones date to 10,705 ± 35 (14)C years bp (approximately 12,707-12,556 calendar years bp) and were directly associated with Clovis tools. We sequenced the genome to an average depth of 14.4× and show that the gene flow from the Siberian Upper Palaeolithic Mal'ta population into Native American ancestors is also shared by the Anzick-1 individual and thus happened before 12,600 years bp. We also show that the Anzick-1 individual is more closely related to all indigenous American populations than to any other group. Our data are compatible with the hypothesis that Anzick-1 belonged to a population directly ancestral to many contemporary Native Americans. Finally, we find evidence of a deep divergence in Native American populations that predates the Anzick-1 individual.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
Extended Data Figure 1
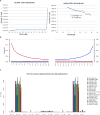
DNA fragmentation, damage and type specific error. a, Left, fragment length distribution of the Anzick-1 DNA sequences mapping to a human reference genome. The maximum read length with the applied chemistry on the HiSeq Illumina platform is 94 bp (100 - 6 bp index read), hence the large peak at this length simply represent the entire tail of the distribution. Right, the declining part of the distribution for the nuclear DNA, and the fit to an exponential model. The decay constant (λ) is estimated to 0.018. b, Damage patterns for the Anzick-1 individual in a random 0.5% subset of all mapped reads. Mismatch frequency relative to the reference as function of read position, C to T in red and G to A in blue. c) Type specific error rates for the Anzick-1 sample and the individual libraries. Estimates of overall error rates are given in the right-hand side.
Extended Data Figure 2
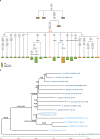
mtDNA and Y-chromosome subtrees. a, Schematic phylogenetic tree of mtDNA haplogroup D4h3 and its sub-branch D4h3a. Mutations from the root of haplogroup D4h are specified only for haplogroup D4h3a lineage, in case of broken lines only the mutations defining an existing sub-branch has been shown. The haplotypes of Anzick-1, identical with the root haplotype of D4h3a, and ancient full sequence from North-western coast of North America (Ancient939) is indicated in red. Insertions are indicated with “.” followed by a number of inserted nucleotides (X if not specified), deletions are indicated with „d“ and back mutations to ancestral state with „!“. The geographical spread of sub-branches of hg D4h is shown with different colours specified in figure legend. b, Placement of Anzick-1 within the Y-chromosome phylogeny. Anzick-1 (circled) represents Y-chromosome haplogroup Q-L54*(xM3) (blue), which is offset by haplogroup Q-M3 (dark blue). The lineage carried by the ancient Saqqaq Palaeo-Eskimo (light blue) constitutes an outgroup to Q-L54. Each branch is labelled by an index and the number of transversion SNPs assigned to the branch (in brackets). Terminal taxa (individuals) are also labelled by population, ID, and haplogroup. Branches 21 and 25 represent the most recent shared ancestry between Anzick-1 and other members of the sample. Branch 19 is significantly shorter than neighbouring branches, which have had an additional ~12,600 years to accumulate mutations.
Extended Data Figure 3
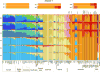
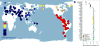
Ancestry proportions of Anzick-1 as determined by ADMIXTURE assuming 3 to 5 and 9 to 11 hypothetical “ancestral” populations or genetic components for a set of 135 extant Eurasian, Oceanian and New World populations. Shown are results from one of the converged runs at each K. We note that the model at K=11 was found to have the best predictive accuracy as determined by the lowest cross validation index values (see SI text). At each K each sample is represented by a stacked vertical bar whereas these of the Anzick-1 are magnified and presented horizontally at the top. Note that irrespective of the number of genetic components assumed the Anzick-1 sample shares all the components present in different contemporary Native American populations.
Extended Data Figure 4
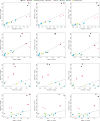
The closer relationship between Anzick-1 and Southern Native Americans compared to Algonquin, Cree, Ojibwa and a Yaqui individual (a–d) is consistent for different 44 Southern and Central Native American populations to Anzick-1. We used the test D(Han, Anzick-1; Algonquin/Cree/Ojibwa/Yaqui, Central/Southern Native Americans). Thick and thin whiskers represent 1 and 3 SEs, respectively.
Extended Data Figure 5
Outgroup _f3_-statistics contrasted for different combinations of populations. (A) Shared genetic history with Anzick-1 compared to shared genetic history with the three Northern Amerind-speaking populations. (B and C) shared genetic history with the Anzick-1 individual compared to the ~4,000-year-old Saqqaq from Greenland. (D and E) Anzick-1 compared to shared genetic history with the 24,000-year-old MA-1 individual from Central Siberia. (F and G) shared genetic history with Anzick-1 compared to shared genetic history with the 40,000 year old Tianyuan individual from China.
Extended Data Figure 6
Pairwise outgroup f3 statistics computed using Saqqaq, Han, French or ancient MA-1 (Mal’ta) on the x-axis and Anzick-1, Karitiana, or Mayan on the y-axis. The black line indicates the y=x line.
Figure 1
Geographic and C14 dating overview, and examples of artefacts from the site. a, Location of the Anzick site relative to continental glacial positions from 16,000 to 13,000 calendar years before present. b, Photograph of the Anzick site. Site is located at the base of the slope at the far left. c, Age of the human remains and osseous tools relative to other Clovis sites. d, Clovis fluted projectile point from the site. e, Clovis osseous rod from the site.
Figure 2
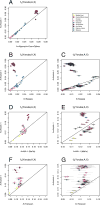
Genetic affinity of Anzick-1. a, Anzick-1 is most closely related to Native Americans. Heat map representing estimated outgroup _f_3-statistics for shared genetic history between the Anzick-1 individual and each of 143 contemporary human populations outside sub-Saharan Africa. b, Anzick-1 is less closely related to Northern Native American populations and a Yaqui individual than to Central and South Native Americans such as the Brazilian Karitiana. We computed a _D_-test of the form D(Han, Anzick-1; Karitiana, X) to test the hypothesis that a second Native American population X is as closely related to Anzick-1 as the South American Karitiana is. Thick and thin whiskers represent 1 and 3 SEs, respectively.
Figure 3
Simplified schematic of genetic models. Alternative models of the population history behind the closer shared ancestry of the Anzick-1 individual to Central and Southern American (SA) populations than Northern American (NA) populations, see main text for further definition of populations. We find that the data is consistent with a simple tree-like model where NA populations are historically basal to Anzick-1 and SA. We base this conclusion on two _D_-tests conducted on the Anzick-1 individual, NA and SA. We used Han Chinese as outgroup. We first tested the hypothesis that Anzick-1 is basal to both NA and SA populations using D(Han, Anzick-1; SA, NA). a, As in the results for each pairwise comparison between SA and NA populations (Extended Data Figure 4), this hypothesis is rejected. b, Next, we tested D(Han, NA; Anzick-1, SA), if NA populations where a mixture of post-Anzick-1 and pre-Anzick-1 ancestry, we would expected to reject this topology. c, We found that a topology with NA populations are basal to Anzick-1 and SA populations is consistent with the data. d, However, another alternative is that the Anzick-1 individual is from the time of the last common ancestral population of the Northern and Southern lineage, after which the Northern lineage received gene flow from a more basal lineage.
Figure 4
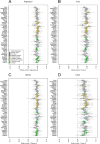
Estimated drift and maximum likelihood tree. Site patterns and drift estimates for non-African populations vs. the Anzick-1 sample. a, Data based on conditioning on African polymorphism, non-African populations in colour on the left, Anzick-1 in grey on the right. b, Data based on conditioning on African polymorphism and removing sites where a C and T or G and A were observed. c, Maximum likelihood tree generated by TreeMix using the whole genome sequencing data with the Mayan genome masked for European ancestry.
Comment in
- Palaeogenomics: genetic roots of the first Americans.
Raff JA, Bolnick DA. Raff JA, et al. Nature. 2014 Feb 13;506(7487):162-3. doi: 10.1038/506162a. Nature. 2014. PMID: 24522593 No abstract available.
Similar articles
- Upper Palaeolithic Siberian genome reveals dual ancestry of Native Americans.
Raghavan M, Skoglund P, Graf KE, Metspalu M, Albrechtsen A, Moltke I, Rasmussen S, Stafford TW Jr, Orlando L, Metspalu E, Karmin M, Tambets K, Rootsi S, Mägi R, Campos PF, Balanovska E, Balanovsky O, Khusnutdinova E, Litvinov S, Osipova LP, Fedorova SA, Voevoda MI, DeGiorgio M, Sicheritz-Ponten T, Brunak S, Demeshchenko S, Kivisild T, Villems R, Nielsen R, Jakobsson M, Willerslev E. Raghavan M, et al. Nature. 2014 Jan 2;505(7481):87-91. doi: 10.1038/nature12736. Epub 2013 Nov 20. Nature. 2014. PMID: 24256729 Free PMC article. - Reassessing the chronology of the archaeological site of Anzick.
Becerra-Valdivia L, Waters MR, Stafford TW Jr, Anzick SL, Comeskey D, Devièse T, Higham T. Becerra-Valdivia L, et al. Proc Natl Acad Sci U S A. 2018 Jul 3;115(27):7000-7003. doi: 10.1073/pnas.1803624115. Epub 2018 Jun 18. Proc Natl Acad Sci U S A. 2018. PMID: 29915063 Free PMC article. - Late Pleistocene exploration and settlement of the Americas by modern humans.
Waters MR. Waters MR. Science. 2019 Jul 12;365(6449):eaat5447. doi: 10.1126/science.aat5447. Science. 2019. PMID: 31296740 Review. - Mitochondrial DNA and Y chromosome diversity and the peopling of the Americas: evolutionary and demographic evidence.
Schurr TG, Sherry ST. Schurr TG, et al. Am J Hum Biol. 2004 Jul-Aug;16(4):420-39. doi: 10.1002/ajhb.20041. Am J Hum Biol. 2004. PMID: 15214060 - The late Pleistocene dispersal of modern humans in the Americas.
Goebel T, Waters MR, O'Rourke DH. Goebel T, et al. Science. 2008 Mar 14;319(5869):1497-502. doi: 10.1126/science.1153569. Science. 2008. PMID: 18339930 Review.
Cited by
- The rise and transformation of Bronze Age pastoralists in the Caucasus.
Ghalichi A, Reinhold S, Rohrlach AB, Kalmykov AA, Childebayeva A, Yu H, Aron F, Semerau L, Bastert-Lamprichs K, Belinskiy AB, Berezina NY, Berezin YB, Broomandkhoshbacht N, Buzhilova AP, Erlikh VR, Fehren-Schmitz L, Gambashidze I, Kantorovich AR, Kolesnichenko KB, Lordkipanidze D, Magomedov RG, Malek-Custodis K, Mariaschk D, Maslov VE, Mkrtchyan L, Nagler A, Fazeli Nashli H, Ochir M, Piotrovskiy YY, Saribekyan M, Sheremetev AG, Stöllner T, Thomalsky J, Vardanyan B, Posth C, Krause J, Warinner C, Hansen S, Haak W. Ghalichi A, et al. Nature. 2024 Nov;635(8040):917-925. doi: 10.1038/s41586-024-08113-5. Epub 2024 Oct 30. Nature. 2024. PMID: 39478221 Free PMC article. - Petrous bones versus tooth cementum for genetic analysis of aged skeletal remains.
Zupanič Pajnič I, Jeromelj T, Leskovar T. Zupanič Pajnič I, et al. Int J Legal Med. 2024 Oct 12. doi: 10.1007/s00414-024-03346-5. Online ahead of print. Int J Legal Med. 2024. PMID: 39394478 - Ancient Rapanui genomes reveal resilience and pre-European contact with the Americas.
Moreno-Mayar JV, Sousa da Mota B, Higham T, Klemm S, Gorman Edmunds M, Stenderup J, Iraeta-Orbegozo M, Laborde V, Heyer E, Torres Hochstetter F, Friess M, Allentoft ME, Schroeder H, Delaneau O, Malaspinas AS. Moreno-Mayar JV, et al. Nature. 2024 Sep;633(8029):389-397. doi: 10.1038/s41586-024-07881-4. Epub 2024 Sep 11. Nature. 2024. PMID: 39261618 Free PMC article. - Spatiotemporal fluctuations of population structure in the Americas revealed by a meta-analysis of the first decade of archaeogenomes.
Dos Santos ALC, Sullasi HSL, Gokcumen O, Lindo J, DeGiorgio M. Dos Santos ALC, et al. Am J Biol Anthropol. 2023 Apr;180(4):703-714. doi: 10.1002/ajpa.24673. Epub 2022 Dec 4. Am J Biol Anthropol. 2023. PMID: 39081397 Free PMC article. - Neandertal ancestry through time: Insights from genomes of ancient and present-day humans.
Iasi LNM, Chintalapati M, Skov L, Mesa AB, Hajdinjak M, Peter BM, Moorjani P. Iasi LNM, et al. bioRxiv [Preprint]. 2024 May 13:2024.05.13.593955. doi: 10.1101/2024.05.13.593955. bioRxiv. 2024. PMID: 38798350 Free PMC article. Updated. Preprint.
References
- Waters MR, Stafford TW. Redefining the age of Clovis: implications for the peopling of the Americas. Science. 2007;315:1122–1126. - PubMed
- Goebel T, Waters MR, O’Rourke DH. The late Pleistocene dispersal of modern humans in the Americas. Science. 2008;319:1497–1502. - PubMed
- Meltzer DJ. First Peoples in a New World. Univ of California Press; 2009.
- Stanford DJ, Bradley BA. Across Atlantic Ice: The Origin of America’s Clovis Culture. University of California Press; 2012.
Publication types
MeSH terms
Substances
Grants and funding
- BB/H008802/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- T15 LM007033/LM/NLM NIH HHS/United States
- BB/H005854/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- P25032/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials