Transcriptome-based network analysis reveals a spectrum model of human macrophage activation - PubMed (original) (raw)
. 2014 Feb 20;40(2):274-88.
doi: 10.1016/j.immuni.2014.01.006. Epub 2014 Feb 13.
Susanne V Schmidt 1, Jil Sander 1, Astrid Draffehn 1, Wolfgang Krebs 1, Inga Quester 1, Dominic De Nardo 2, Trupti D Gohel 1, Martina Emde 1, Lisa Schmidleithner 1, Hariharasudan Ganesan 1, Andrea Nino-Castro 1, Michael R Mallmann 1, Larisa Labzin 2, Heidi Theis 1, Michael Kraut 1, Marc Beyer 1, Eicke Latz 3, Tom C Freeman 4, Thomas Ulas 1, Joachim L Schultze 5
Affiliations
- PMID: 24530056
- PMCID: PMC3991396
- DOI: 10.1016/j.immuni.2014.01.006
Transcriptome-based network analysis reveals a spectrum model of human macrophage activation
Jia Xue et al. Immunity. 2014.
Abstract
Macrophage activation is associated with profound transcriptional reprogramming. Although much progress has been made in the understanding of macrophage activation, polarization, and function, the transcriptional programs regulating these processes remain poorly characterized. We stimulated human macrophages with diverse activation signals, acquiring a data set of 299 macrophage transcriptomes. Analysis of this data set revealed a spectrum of macrophage activation states extending the current M1 versus M2-polarization model. Network analyses identified central transcriptional regulators associated with all macrophage activation complemented by regulators related to stimulus-specific programs. Applying these transcriptional programs to human alveolar macrophages from smokers and patients with chronic obstructive pulmonary disease (COPD) revealed an unexpected loss of inflammatory signatures in COPD patients. Finally, by integrating murine data from the ImmGen project we propose a refined, activation-independent core signature for human and murine macrophages. This resource serves as a framework for future research into regulation of macrophage activation in health and disease.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures
Graphical abstract
Figure 1
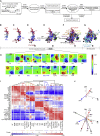
Extending Macrophage Polarization to a Spectrum Model (A) Schema describing the workflow for Figure 1. (B–G) Correlation networks of 299 macrophage transcriptomes representing 29 conditions from end of activation time points (B–F) also including intermediate time points (G). (H) Self-Organizing Map (SOM) clustering using samples displayed in (E). Clusters with the top up- or downregulated genes for each stimulus are marked with a frame. (I) Matrix of hierarchically clustered Pearson’s correlation coefficient matrix (CCM) standardized from −2.78 to 2.78 (blue to red) based on 1,000 most variable probes. (J) Spectrum model (3D) based on the ten clusters defined in (I) and sample values (coordinates) defined by correlation network from (F). Baseline macrophages (Mb) are set as origin, activation states are represented by colored arrows, x, y, z axes are in dashed lines with double arrows. See also Figure S1, Movie S1, and Table S2.
Figure 2
Genes with Selective Expression Associated with Distinct Stimuli (A) Schema describing the workflow for Figure 2. (B) Absolute expression counts (mean ± SD) of genes defined by SOM clustering to be highly expressed for a particular stimulation condition. Shown here are genes enriched in either Mb (baseline), IFN-β, GC, IL4+uLPS, upLPS+IC, PA, IFN-γ, IFN-γ +TNF, sLPS+IFN-γ, or TPP+IFN-β. All other conditions are shown in Figure S2.
Figure 3
Activation-Specific Genes Revealed by Weighted Correlation Network Analysis (A) Schema describing the workflow for Figure 3. (B) Heatmap showing the correlation of the module eigengene (first principal component; ME) to the traits (stimulation conditions). Blue means negative correlation; orange means positive correlation. (C) Network visualization of GOEA of modules 30, 32–33 (positively correlated), respectively, 12, 13, and 20 (negatively correlated) for TPP-stimulation using BiNGO and EnrichmentMap. Red nodes represent enriched GO-terms, whereas node size represents corresponding FDR-adjusted enrichment p value (q value). Edge thickness shows overlap of genes between neighbor nodes. (D) Correlation network of module-specific transcription factors (TFs) (IFN-γ: modules 7-9; IL-4: modules 13-15; TPP: 30, 32–33). Edge width shows the Pearson’s correlation between each TF pair. See also Figure S3 and Table S2.
Figure 4
Phenotypic and Functional Characterization of Macrophages Stimulated with TNF, PGE2, and P3C (A) Flow cytometry of CD14, CD23, CD25, CD86, CXCR7, CD197, in Mb (dark gray), IFN-γ (light blue), IL-4 (red), and TPP (light gray) activated macrophages. Mean fluorescence intensities (MFI) of at least three independent experiments are shown (mean and SEM; p∗ < 0.05 Student’s t test). (B) Immunoblot analysis of STAT4 and β-actin. STAT4 expression was normalized to β-actin expression and set to 100% in MTPP (TPP). (C) CXCL5 and IL1α in supernatants of macrophage cultures. (D) T cell activation by CD3 and CD28 beads in presence or absence of macrophages assessed by CFSE dilution. (E) Heatmap showing fold changes of highly abundant miRNAs up- or downregulated (FC > 2, FDR adjusted p value < 0.05) in M1 (IFN-γ) or M2 (IL-4), or MTPP (TPP) compared to Mb (baseline). Fold changes colored from blue to red. For Figures 4A–4D, mean ± SEM (p∗ < 0.05, Student’s t-test) are shown.
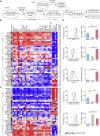
Figure 5
Enrichment of Activation Modules in Human Alveolar Macrophages (A) Schema describing the workflow for Figure 5. (B) Correlation network of human alveolar macrophages (n = 100) from two studies (Shaykhiev et al., 2009; Woodruff et al., 2005) using 374 genes differentially expressed between nonsmokers (n = 39) and smokers (n = 49) or COPD (n = 12) patients (FC > 2.0, FDR adjusted p value < 0.05). (C) Volcano plots of normalized enrichment scores (NES) and enrichment p values based on GSEA using WGCNA modules defined in Figure 3. Shown are data for the stimuli IFN-γ, IL-4, TPP, and palmitic acid (PA). Red circles show gene sets positively significantly enriched (NES > 1, p value < 0.05); blue circles show gene sets significantly depleted (NES < −1, p value < 0.05). (D) Volcano plots of normalized enrichment scores (NES) and enrichment p values of the same gene sets applied to data from alveolar macrophages derived from smokers and COPD patients. Representative results of several permutation runs of GSEA. (E) Network visualization of GOEA of positively enriched modules (p value < 0.01) for smokers (modules 41, 6, 21, 16, 39, 5, 19, and 42) and negatively enriched modules (p value < 0.01) for COPD patients (modules 8, 29) using BiNGO and EnrichmentMap. Red nodes represent enriched GO-terms, node size, and color represent corresponding FDR-adjusted enrichment p values (q values).
Figure 6
Macrophage Activation Network Calculated by ARACNe (A) Schema describing the workflow for Figure 6. (B) Visualization of the 10% largest hub genes of the ARACNe predicted macrophage regulatory network (n = 299 transcriptomes). For the top ten genes (highest degree of connectivity, blue) and TFs, mean expression values (log2, derived from the ten clusters in Figure 1I) are highlighted in red colors. Node size reflects degree of connectivity. (C) Network visualization of GOEA using BiNGO and EnrichmentMap on hubs shown in (B). Red nodes represent enriched GO-terms, and node size represent FDR-adjusted enrichment p value (q value). Edge thickness represents overlap of genes between neighbor nodes. (D) Subnetworks of the five most highly expressed TFs from all hubs shown in (B). First neighbors are surrounding corresponding TFs. Each gene is multicolored according to its mean expression (log2) in ten clusters (from Figure 1I). (E) Top 30 putative candidates after Gene Prioritization (GP) of 869 hubs. Mean expression (log2) from each cluster is displayed as a heatmap. Categorization is according to cellular functions. See also Figures S4 and S5 and Table S3.
Figure 7
Expression of murine macrophage and dendritic cell signature genes in humans (A) Schema describing the workflow for Figure 7. M, macrophage. (B and C) Heatmap (standardized and scaled log2 expression) of human orthologs of murine (B) macrophage signature genes (Gautier et al., 2012) and (C) DCs signature genes (Miller et al., 2012) in monocytes, 29 macrophage stimulation conditions, and monocyte-derived DCs (baseline = GM-CSF+IL4). TPP = TNF+PGE2+P3C. (D) Flow cytometric analysis for surface markers CD14, CD32, MERTK, CD64, CD13 on human monocytes (Mono, blue), baseline DCs (DCb, green), and baseline macrophages (Mb, red). Mean fluorescence intensities (MFI) of at least three independent experiments (mean and SEM; p∗ < 0.05 Student’s t test). See also Table S4.
Comment in
- Macrophage activation: glancing into diversity.
Natoli G, Monticelli S. Natoli G, et al. Immunity. 2014 Feb 20;40(2):175-7. doi: 10.1016/j.immuni.2014.01.004. Immunity. 2014. PMID: 24560195
Similar articles
- Transcriptional and functional diversity of human macrophage repolarization.
Gharib SA, McMahan RS, Eddy WE, Long ME, Parks WC, Aitken ML, Manicone AM. Gharib SA, et al. J Allergy Clin Immunol. 2019 Apr;143(4):1536-1548. doi: 10.1016/j.jaci.2018.10.046. Epub 2018 Nov 14. J Allergy Clin Immunol. 2019. PMID: 30445062 Free PMC article. - Control of the Inflammatory Macrophage Transcriptional Signature by miR-155.
Jablonski KA, Gaudet AD, Amici SA, Popovich PG, Guerau-de-Arellano M. Jablonski KA, et al. PLoS One. 2016 Jul 22;11(7):e0159724. doi: 10.1371/journal.pone.0159724. eCollection 2016. PLoS One. 2016. PMID: 27447824 Free PMC article. - Transcriptome analysis of long non-coding RNAs in Mycobacterium avium complex-infected macrophages.
Yoshida M, Kwon AT, Qin XY, Nishimura H, Maeda S, Miyamoto Y, Yoshida Y, Hoshino Y, Suzuki H. Yoshida M, et al. Front Immunol. 2024 Apr 22;15:1374437. doi: 10.3389/fimmu.2024.1374437. eCollection 2024. Front Immunol. 2024. PMID: 38711507 Free PMC article. - MiRNA-Mediated Macrophage Polarization and its Potential Role in the Regulation of Inflammatory Response.
Essandoh K, Li Y, Huo J, Fan GC. Essandoh K, et al. Shock. 2016 Aug;46(2):122-31. doi: 10.1097/SHK.0000000000000604. Shock. 2016. PMID: 26954942 Free PMC article. Review. - Roles of Macrophage Polarization and Macrophage-Derived miRNAs in Pulmonary Fibrosis.
Kishore A, Petrek M. Kishore A, et al. Front Immunol. 2021 Aug 13;12:678457. doi: 10.3389/fimmu.2021.678457. eCollection 2021. Front Immunol. 2021. PMID: 34489932 Free PMC article. Review.
Cited by
- Susceptibility to Intracellular Infections: Contributions of TNF to Immune Defense.
Li X, Körner H, Liu X. Li X, et al. Front Microbiol. 2020 Jul 15;11:1643. doi: 10.3389/fmicb.2020.01643. eCollection 2020. Front Microbiol. 2020. PMID: 32760383 Free PMC article. Review. - Intestinal Inflammation Modulates the Expression of ACE2 and TMPRSS2 and Potentially Overlaps With the Pathogenesis of SARS-CoV-2-related Disease.
Suárez-Fariñas M, Tokuyama M, Wei G, Huang R, Livanos A, Jha D, Levescot A, Irizar H, Kosoy R, Cording S, Wang W, Losic B, Ungaro RC, Di'Narzo A, Martinez-Delgado G, Suprun M, Corley MJ, Stojmirovic A, Houten SM, Peters L, Curran M, Brodmerkel C, Perrigoue J, Friedman JR, Hao K, Schadt EE, Zhu J, Ko HM, Cho J, Dubinsky MC, Sands BE, Ndhlovu L, Cerf-Bensusan N, Kasarskis A, Colombel JF, Harpaz N, Argmann C, Mehandru S. Suárez-Fariñas M, et al. Gastroenterology. 2021 Jan;160(1):287-301.e20. doi: 10.1053/j.gastro.2020.09.029. Epub 2020 Sep 25. Gastroenterology. 2021. PMID: 32980345 Free PMC article. - Immunosuppression in Sepsis: Biomarkers and Specialized Pro-Resolving Mediators.
Padovani CM, Yin K. Padovani CM, et al. Biomedicines. 2024 Jan 13;12(1):175. doi: 10.3390/biomedicines12010175. Biomedicines. 2024. PMID: 38255280 Free PMC article. Review. - A data-driven computational model enables integrative and mechanistic characterization of dynamic macrophage polarization.
Zhao C, Medeiros TX, Sové RJ, Annex BH, Popel AS. Zhao C, et al. iScience. 2021 Jan 29;24(2):102112. doi: 10.1016/j.isci.2021.102112. eCollection 2021 Feb 19. iScience. 2021. PMID: 33659877 Free PMC article. - GlioM&M: Web-based tool for studying circulating and infiltrating monocytes and macrophages in glioma.
Abels ER, Maas SLN, Tai E, Ting DT, Broekman MLD, Breakefield XO, El Khoury J. Abels ER, et al. Sci Rep. 2020 Jun 18;10(1):9898. doi: 10.1038/s41598-020-66728-w. Sci Rep. 2020. PMID: 32555306 Free PMC article.
References
- Barnes P.J. New anti-inflammatory targets for chronic obstructive pulmonary disease. Nat. Rev. Drug Discov. 2013;12:543–559. - PubMed
- Basso K., Margolin A.A., Stolovitzky G., Klein U., Dalla-Favera R., Califano A. Reverse engineering of regulatory networks in human B cells. Nat. Genet. 2005;37:382–390. - PubMed
Publication types
MeSH terms
Grants and funding
- BB/F003722/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/I001107/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/D/05191131/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/D/20211552/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases