A genetic atlas of human admixture history - PubMed (original) (raw)
A genetic atlas of human admixture history
Garrett Hellenthal et al. Science. 2014.
Abstract
Modern genetic data combined with appropriate statistical methods have the potential to contribute substantially to our understanding of human history. We have developed an approach that exploits the genomic structure of admixed populations to date and characterize historical mixture events at fine scales. We used this to produce an atlas of worldwide human admixture history, constructed by using genetic data alone and encompassing over 100 events occurring over the past 4000 years. We identified events whose dates and participants suggest they describe genetic impacts of the Mongol empire, Arab slave trade, Bantu expansion, first millennium CE migrations in Eastern Europe, and European colonialism, as well as unrecorded events, revealing admixture to be an almost universal force shaping human populations.
Figures
Fig. 1. Ancestry painting and admixture analysis of simulated admixture
(A) We illustrate a simulated event 30 generations ago between Brahui (80%, red) and Yoruba (20%, yellow), resulting in admixed individuals having DNA segments from each source (bottom). The true sources are then treated as unsampled (B) CHROMOPAINTER’s painting of the same region (yellow=Africa, green=America, red=Central-South-Asia, blue=East-Asia, cyan=Europe, pink=Near-East, black=Oceania), showing haplotypic segments (“chunks”) shared with these groups. Our model fitting narrows the donor set largely to Central-South Asia and Africa, generating a “cleaned” painting. (C) Coancestry curves (black line) show relative probability of jointly copying two chunks from red (Balochi; FST =0.003 with Brahui) and/or yellow (Mandenka; FST =0.009 with Yoruba) donors, at varying genetic distances. The curves closely fit an exponential decay (green line) with rate 30 generations (95% CI: 27-33). The positive slope for the Balochi – Mandenka curve (middle) implies these donors represent different admixing sources. (D) GLOBETROTTER’s source inference, with black diamonds indicating sampled populations with greatest similarity (_FST_≤0.001 over minimum) to true sources, white circles other sampled populations. Red and yellow circles, with areas summing to 20% and 80%, respectively, show inferred haplotypic make-up of the two admixing sources.
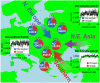
Fig. 2. Overview of inferred admixture for 95 human populations
(A) Coancestry curve for the Maya for Spanish donor group (inferred as closest to minor admixing source), with green fitted line showing inferred exponential decay curve and a corresponding recent admixture date (with 95% CI). (B, C) As A, but showing the Druze and Kalash respectively, with different indicated donors (donors indicated are proxies for minor admixing source, inferred as closest to Yoruba and Germany/Austria, respectively) and with successively older admixture. (D) On the map (locations approximate in densely sampled regions), shapes (see legend) indicate inference: no admixture, a single admixture event, or more complex admixture; colors indicate fineSTRUCTURE clustering into 18 clades (Table S11, Figs. S12-S13). Inferred date(s), 95% CIs are directly below the map, with two inferred admixing sources (dots and vertical bars) shown below each date (see example for simulation of Fig. 1 at left). For multiple admixture times, these two sources correspond to the more recent event; for multiple groups, they reflect the strongest admixture “direction”. Colored dots above each bar indicate clades best representing the major (top) and minor (bottom) sources. The bar is split at the inferred admixture fraction (horizontal line, fractions < 5% shown as 5%). Each bar section indicates inferred donor group haplotypic make-up, colored as the map, for one source. Shaded boxes on the inferred admixture times denote events referred to in the text, specifically 1. European colonization of the Americas (1492CE-present; hot pink), 2. Slavic (500-900CE; pink), Turkic (500-1100CE; maroon) migrations, 3. Arab slave trade (650-1900CE; cyan), 4. Mongol Empire (1206-1368CE; purple), and 5. Khmer Empire (802-1431CE; orange).
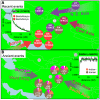
Fig. 3. Multi-way admixture in Eastern Europe
Mixing percentages (pie graphs) and dates (white text) inferred using the strongest admixture “direction” for 6 eastern European groups: Belarus (BE), Bulgaria (BU), Hungary (HU), Lithuania (LI), Poland (PO), Romania (RO), analyzed when disallowing copying from nearby groups, and Greece (GR), analyzed using the full set of 94 donors. Mixing percentages indicate percentages for three geographic regions: “N. Europe” (Northwest Europe and East Europe from clades of Table S11; blue), “Southern” (South Europe and West Asia; red) and “N.E.Asia” (Northeast Asia and Yakut; purple, also given above each pie), plus “other” (grey). All groups except Greece show evidence (p<0.05) of multi-way admixture involving sources along the approximate directions show by the arrows. Coancestry curves (black lines) for Bulgaria, fitted with an exponential decay curve (green lines), exemplify this multi-way signal. Each pairing of the three donor groups, each a proxy for the admixture source from a different region (Norway: N/E Europe, Oroqen: NE Asia, Greece: S. Europe and W. Asia), exhibits negative correlation (a dip) in ancestry weights at short genetic distances, implying at least three identifiably distinct ancestral sources mixing (approximately) simultaneously (9).
Fig. 4. Ancient and modern admixture in Central Asia
(A) Dates (white text) and minority contributing sources for recent inferred events in 9 populations (circles), analyzed disallowing copying from nearby groups, showing contributions from Northeast Asia (purple) in the Hazara (HA), Uygur (UY) and Uzbekistani (UZ); East Asia (maroon) in Burusho (BU); West Asia (brown) in Pathan (PA); Africa (red) in Balochi (BA), Brahui (BR), Makrani (MA) and Sindhi (SI). Kalash (KA, grey) have no inferred recent event. (B) Inferred mixing percentages (pie graphs) and dates (white text gives upper CI bound) for additional, possibly shared, ancient events in 7 groups. Pie graphs show inferred donor make-up of each group after removing the recent event contribution from (A), if any, with colors referring to donors from “East Asia” (Southeast Asia from clades of Table S11; maroon), “Europe” (Northwest, East and South Europe; hot pink), Central “South Asia” (orange), “West Asia” (brown) and “other” (white). Arrows indicate “directions” of ancient admixture, with donor regions splitting into two pairs representing different sources. Coancestry curves (black lines) for Sindhi are superimposed for two different donor pairs representing proxies for admixing groups with ancestry indicated by the filled circles, indicating highly different exponential decay rates fit as a mixture of 7 and 94 generations (green lines).
Similar articles
- A Genetic History of the Near East from an aDNA Time Course Sampling Eight Points in the Past 4,000 Years.
Haber M, Nassar J, Almarri MA, Saupe T, Saag L, Griffith SJ, Doumet-Serhal C, Chanteau J, Saghieh-Beydoun M, Xue Y, Scheib CL, Tyler-Smith C. Haber M, et al. Am J Hum Genet. 2020 Jul 2;107(1):149-157. doi: 10.1016/j.ajhg.2020.05.008. Epub 2020 May 28. Am J Hum Genet. 2020. PMID: 32470374 Free PMC article. - The genetic legacy of the expansion of Turkic-speaking nomads across Eurasia.
Yunusbayev B, Metspalu M, Metspalu E, Valeev A, Litvinov S, Valiev R, Akhmetova V, Balanovska E, Balanovsky O, Turdikulova S, Dalimova D, Nymadawa P, Bahmanimehr A, Sahakyan H, Tambets K, Fedorova S, Barashkov N, Khidiyatova I, Mihailov E, Khusainova R, Damba L, Derenko M, Malyarchuk B, Osipova L, Voevoda M, Yepiskoposyan L, Kivisild T, Khusnutdinova E, Villems R. Yunusbayev B, et al. PLoS Genet. 2015 Apr 21;11(4):e1005068. doi: 10.1371/journal.pgen.1005068. eCollection 2015 Apr. PLoS Genet. 2015. PMID: 25898006 Free PMC article. - Bantu-speaker migration and admixture in southern Africa.
Choudhury A, Sengupta D, Ramsay M, Schlebusch C. Choudhury A, et al. Hum Mol Genet. 2021 Apr 26;30(R1):R56-R63. doi: 10.1093/hmg/ddaa274. Hum Mol Genet. 2021. PMID: 33367711 Free PMC article. Review. - Chad Genetic Diversity Reveals an African History Marked by Multiple Holocene Eurasian Migrations.
Haber M, Mezzavilla M, Bergström A, Prado-Martinez J, Hallast P, Saif-Ali R, Al-Habori M, Dedoussis G, Zeggini E, Blue-Smith J, Wells RS, Xue Y, Zalloua PA, Tyler-Smith C. Haber M, et al. Am J Hum Genet. 2016 Dec 1;99(6):1316-1324. doi: 10.1016/j.ajhg.2016.10.012. Epub 2016 Nov 23. Am J Hum Genet. 2016. PMID: 27889059 Free PMC article. - Admixture and Ancestry Inference from Ancient and Modern Samples through Measures of Population Genetic Drift.
Harris AM, DeGiorgio M. Harris AM, et al. Hum Biol. 2017 Jan;89(1):21-46. doi: 10.13110/humanbiology.89.1.02. Hum Biol. 2017. PMID: 29285965 Review.
Cited by
- The genetic history of Greenlandic-European contact.
Waples RK, Hauptmann AL, Seiding I, Jørsboe E, Jørgensen ME, Grarup N, Andersen MK, Larsen CVL, Bjerregaard P, Hellenthal G, Hansen T, Albrechtsen A, Moltke I. Waples RK, et al. Curr Biol. 2021 May 24;31(10):2214-2219.e4. doi: 10.1016/j.cub.2021.02.041. Epub 2021 Mar 11. Curr Biol. 2021. PMID: 33711251 Free PMC article. - The Spatial Mixing of Genomes in Secondary Contact Zones.
Sedghifar A, Brandvain Y, Ralph P, Coop G. Sedghifar A, et al. Genetics. 2015 Sep;201(1):243-61. doi: 10.1534/genetics.115.179838. Epub 2015 Jul 22. Genetics. 2015. PMID: 26205988 Free PMC article. - The impact of recent population history on the deleterious mutation load in humans and close evolutionary relatives.
Simons YB, Sella G. Simons YB, et al. Curr Opin Genet Dev. 2016 Dec;41:150-158. doi: 10.1016/j.gde.2016.09.006. Epub 2016 Oct 13. Curr Opin Genet Dev. 2016. PMID: 27744216 Free PMC article. Review. - A genetic method for dating ancient genomes provides a direct estimate of human generation interval in the last 45,000 years.
Moorjani P, Sankararaman S, Fu Q, Przeworski M, Patterson N, Reich D. Moorjani P, et al. Proc Natl Acad Sci U S A. 2016 May 17;113(20):5652-7. doi: 10.1073/pnas.1514696113. Epub 2016 May 2. Proc Natl Acad Sci U S A. 2016. PMID: 27140627 Free PMC article. - Analysis of Gyimes Csango population samples on a high-resolution genome-wide basis.
Bánfai Z, Büki G, Ádám V, Sümegi K, Szabó A, Hadzsiev K, Erős K, Gallyas F, Miseta A, Kásler M, Melegh B. Bánfai Z, et al. BMC Genomics. 2024 Oct 7;25(1):942. doi: 10.1186/s12864-024-10833-x. BMC Genomics. 2024. PMID: 39375616 Free PMC article.
References
- Baird SJE. Fisher’s markers of admixture. Heredity. 2006;97:81–83. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 098387/WT_/Wellcome Trust/United Kingdom
- 098386/Z/12/Z/WT_/Wellcome Trust/United Kingdom
- BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- 090532/WT_/Wellcome Trust/United Kingdom
- 098387/Z/12/Z/WT_/Wellcome Trust/United Kingdom
- WT_/Wellcome Trust/United Kingdom
- 098386/WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous