Structural motif of polyglutamine amyloid fibrils discerned with mixed-isotope infrared spectroscopy - PubMed (original) (raw)
Structural motif of polyglutamine amyloid fibrils discerned with mixed-isotope infrared spectroscopy
Lauren E Buchanan et al. Proc Natl Acad Sci U S A. 2014.
Abstract
Polyglutamine (polyQ) sequences are found in a variety of proteins, and mutational expansion of the polyQ tract is associated with many neurodegenerative diseases. We study the amyloid fibril structure and aggregation kinetics of K2Q24K2W, a model polyQ sequence. Two structures have been proposed for amyloid fibrils formed by polyQ peptides. By forming fibrils composed of both (12)C and (13)C monomers, made possible by protein expression in Escherichia coli, we can restrict vibrational delocalization to measure 2D IR spectra of individual monomers within the fibrils. The spectra are consistent with a β-turn structure in which each monomer forms an antiparallel hairpin and donates two strands to a single β-sheet. Calculated spectra from atomistic molecular-dynamics simulations of the two proposed structures confirm the assignment. No spectroscopically distinct intermediates are observed in rapid-scan 2D IR kinetics measurements, suggesting that aggregation is highly cooperative. Although 2D IR spectroscopy has advantages over linear techniques, the isotope-mixing strategy will also be useful with standard Fourier transform IR spectroscopy.
Keywords: Huntington disease; antiparallel β-sheets; isotope dilution; two-dimensional infrared spectroscopy.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
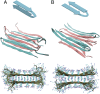
Fig. 1.
Visualizations of fibril models used in MD simulations. Schematic of monomer conformation (Top), one-half of the fibril unit cell (Middle), and fibril cross-section (Bottom) for the (A) β-arc and (B) β-turn fibril models.
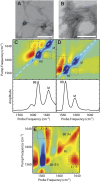
Fig. 2.
Transmission electron microscopy and experimental 2D IR spectra of Q24 fibrils. Electron micrographs of (A) unlabeled and (B) 13C-labeled Q24 fibrils. In each image, the white scale bar represents 200 nm. Two-dimensional IR spectra and diagonal intensity slices of (C) unlabeled and (D) 13C-labeled Q24 fibrils. (E) Polarization difference spectrum of 13C-labeled Q24 fibrils. Diagonal peaks are labeled BB for backbone modes and M for mixed backbone–side-chain modes. Symmetry labels are added for backbone modes in the difference spectrum. Cross-peaks between diagonal modes are highlighted with boxes. Frequencies are summarized in
Table S1
.
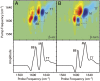
Fig. 3.
Simulated 2D IR spectra of Q24 fibril models. Simulated 2D IR spectra and diagonal intensity slices for the (A) β-arc and (B) β-turn models of unlabeled Q24 fibrils. Diagonal peaks are labeled BB for backbone modes, M for mixed backbone–side-chain modes, and TT for modes from the disordered turns and termini. Cross-peaks between the BB and M modes are highlighted with boxes. Frequencies are summarized in
Table S1
.
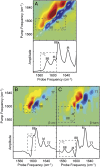
Fig. 4.
Experimental and simulated 2D IR spectra of isotope diluted fibrils. (A) Experimental 2D IR spectrum and diagonal intensity slice of 10% 13C-labeled Q24 fibrils. Simulated 2D IR spectra and diagonal intensity slices for (B) β-arc and (C) β-turn models of ∼8% labeled Q24 fibrils. Diagonal peaks are labeled with BB for backbone modes, M for mixed backbone–side-chain modes, and TT for disordered modes from the turns and termini. Peaks arising from 13C-labeled peptides are highlighted with boxes. Peak frequencies are summarized in
Table S2
.
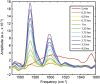
Fig. 5.
Kinetics of Q24 aggregation. Difference slices obtained from 2D IR spectra of 13C-Q24. The initial slice (red) was subtracted from subsequent slices to highlight changes.
Similar articles
- D-polyglutamine amyloid recruits L-polyglutamine monomers and kills cells.
Kar K, Arduini I, Drombosky KW, van der Wel PC, Wetzel R. Kar K, et al. J Mol Biol. 2014 Feb 20;426(4):816-29. doi: 10.1016/j.jmb.2013.11.019. Epub 2013 Nov 28. J Mol Biol. 2014. PMID: 24291210 Free PMC article. - Exploding the Repeat Length Paradigm while Exploring Amyloid Toxicity in Huntington's Disease.
Wetzel R. Wetzel R. Acc Chem Res. 2020 Oct 20;53(10):2347-2357. doi: 10.1021/acs.accounts.0c00450. Epub 2020 Sep 25. Acc Chem Res. 2020. PMID: 32975927 - β-hairpin-mediated nucleation of polyglutamine amyloid formation.
Kar K, Hoop CL, Drombosky KW, Baker MA, Kodali R, Arduini I, van der Wel PC, Horne WS, Wetzel R. Kar K, et al. J Mol Biol. 2013 Apr 12;425(7):1183-97. doi: 10.1016/j.jmb.2013.01.016. Epub 2013 Jan 23. J Mol Biol. 2013. PMID: 23353826 Free PMC article. - Amide I two-dimensional infrared spectroscopy of proteins.
Ganim Z, Chung HS, Smith AW, Deflores LP, Jones KC, Tokmakoff A. Ganim Z, et al. Acc Chem Res. 2008 Mar;41(3):432-41. doi: 10.1021/ar700188n. Epub 2008 Feb 21. Acc Chem Res. 2008. PMID: 18288813 Review. - Protein denaturation and aggregation: Cellular responses to denatured and aggregated proteins.
Meredith SC. Meredith SC. Ann N Y Acad Sci. 2005 Dec;1066:181-221. doi: 10.1196/annals.1363.030. Ann N Y Acad Sci. 2005. PMID: 16533927 Review.
Cited by
- Two-dimensional infrared spectroscopy measures the structural dynamics of a self-assembled film only one molecule thick.
Zanni MT. Zanni MT. Proc Natl Acad Sci U S A. 2016 May 3;113(18):4890-1. doi: 10.1073/pnas.1605263113. Epub 2016 Apr 19. Proc Natl Acad Sci U S A. 2016. PMID: 27095845 Free PMC article. No abstract available. - Conformational studies of pathogenic expanded polyglutamine protein deposits from Huntington's disease.
Matlahov I, van der Wel PC. Matlahov I, et al. Exp Biol Med (Maywood). 2019 Dec;244(17):1584-1595. doi: 10.1177/1535370219856620. Epub 2019 Jun 15. Exp Biol Med (Maywood). 2019. PMID: 31203656 Free PMC article. Review. - Stability of Iowa mutant and wild type Aβ-peptide aggregates.
Alred EJ, Scheele EG, Berhanu WM, Hansmann UH. Alred EJ, et al. J Chem Phys. 2014 Nov 7;141(17):175101. doi: 10.1063/1.4900892. J Chem Phys. 2014. PMID: 25381547 Free PMC article. - Template-assisted design of monomeric polyQ models to unravel the unique role of glutamine side chains in disease-related aggregation.
Siu HW, Heck B, Kovermann M, Hauser K. Siu HW, et al. Chem Sci. 2020 Oct 28;12(1):412-426. doi: 10.1039/d0sc05299j. eCollection 2021 Jan 7. Chem Sci. 2020. PMID: 33552461 Free PMC article. - Vibrational Spectroscopic Map, Vibrational Spectroscopy, and Intermolecular Interaction.
Baiz CR, Błasiak B, Bredenbeck J, Cho M, Choi JH, Corcelli SA, Dijkstra AG, Feng CJ, Garrett-Roe S, Ge NH, Hanson-Heine MWD, Hirst JD, Jansen TLC, Kwac K, Kubarych KJ, Londergan CH, Maekawa H, Reppert M, Saito S, Roy S, Skinner JL, Stock G, Straub JE, Thielges MC, Tominaga K, Tokmakoff A, Torii H, Wang L, Webb LJ, Zanni MT. Baiz CR, et al. Chem Rev. 2020 Aug 12;120(15):7152-7218. doi: 10.1021/acs.chemrev.9b00813. Epub 2020 Jun 29. Chem Rev. 2020. PMID: 32598850 Free PMC article. Review.
References
- Zoghbi HY, Orr HT. Glutamine repeats and neurodegeneration. Annu Rev Neurosci. 2000;23:217–247. - PubMed
- Martin JB. Molecular basis of the neurodegenerative disorders. N Engl J Med. 1999;340(25):1970–1980. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases