A dynamically assembled cell wall synthesis machinery buffers cell growth - PubMed (original) (raw)
A dynamically assembled cell wall synthesis machinery buffers cell growth
Timothy K Lee et al. Proc Natl Acad Sci U S A. 2014.
Abstract
Assembly of protein complexes is a key mechanism for achieving spatial and temporal coordination in processes involving many enzymes. Growth of rod-shaped bacteria is a well-studied example requiring such coordination; expansion of the cell wall is thought to involve coordination of the activity of synthetic enzymes with the cytoskeleton via a stable complex. Here, we use single-molecule tracking to demonstrate that the bacterial actin homolog MreB and the essential cell wall enzyme PBP2 move on timescales orders of magnitude apart, with drastically different characteristic motions. Our observations suggest that PBP2 interacts with the rest of the synthesis machinery through a dynamic cycle of transient association. Consistent with this model, growth is robust to large fluctuations in PBP2 abundance. In contrast to stable complex formation, dynamic association of PBP2 is less dependent on the function of other components of the synthesis machinery, and buffers spatially distributed growth against fluctuations in pathway component concentrations and the presence of defective components. Dynamic association could generally represent an efficient strategy for spatiotemporal coordination of protein activities, especially when excess concentrations of system components are inhibitory to the overall process or deleterious to the cell.
Keywords: Pencillin binding proteins; bacterial cell wall; multienzyme complexes; superresolution microscopy.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
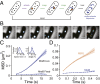
Single-molecule dynamics reveal that the cell wall synthesis enzyme PBP2 undergoes fast, diffusive motion, unlike the directed motion of the MreB cytoskeleton. (A) Schematic of sptPALM. Photoactivatable fluorescent protein fusions are used to monitor the movement of individual proteins in a crowded environment. A small population of molecules is activated by UV laser light, and then imaged over time to track molecular positions. (B) TIRF images of MreBsw-PAmCherry single molecules in live E. coli TKL039 (MG1655 mreB sw -PAmCherry) cells taken every 2.5 s, overlaid on phase-contrast images. The solid lines in the last panel are representative MreB molecular tracks. (Scale bar: 1 μm.) (C) Mean squared displacement (MSD) of MreB molecules in live (n = 382 molecules) and fixed (n = 105) E. coli TKL039 cells. The shaded area represents SEM. (Inset) The distribution of MreB track speeds (28.6 ± 15.4 nm⋅s−1, SD) at 37 °C and angles relative to the cell midline (99.1 ± 29.4°, SD). Most trajectories were approximately perpendicular to the long axis of the cell, with a slight right-handed bias quantitatively consistent with previous studies of left-handed twisting during growth (42). (D) MSD of MreB molecules (n = 1,018) in TKL039 cells and PBP2 molecules (n = 854) in TKL112 (∆mrdA Pmrda-PAmCherry-mrdA) imaged at high frame rates. The shaded area represents SEM. The linear MSD of PBP2 indicates that PBP2 molecules move diffusively, with a diffusion constant of D = 0.06 ± 0.006 µm2/s.
Fig. 2.
Rapid PBP2 diffusion is independent of catalytic activity, and buffers cell growth rate from fluctuations in PBP2 concentration. (A) Representative image of MreB and PBP2 colocalization in a TKL233 (mreB sw -sfGFP ∆mrdA PmrdA-PAmCherry-mrdA) cell filamented with cephalexin. (Upper) MreBsw-sfGFP and PAmCherry-PBP2 were coimaged via TIRF. (Lower) The fluorescence profiles over the length of the cell, averaged over the transverse direction. MreB and PBP2 profiles have a small, but significant correlation [two-sample t test: experimental data vs. scrambled data, t(76) = 2.79, P = 0.0068; Materials and Methods;
Fig. S6
]. (B) The apparent PBP2 diffusion constant is unaffected by antibiotic treatment. SEM is shown (n = 5 experiments). Mecillinam binds the active site of PBP2, A22 disrupts MreB polymerization, and an S330A mutation in PBP2 ablates the catalytic active site. Concentrations are given in micrograms per milliliter. The diffusion constant increases roughly 25-fold when the transpeptidase domain is truncated. (C) Model for PBP2 interaction with MreB. PBP2 moves rapidly within the inner membrane (IM), forming transient interactions with sites of peptidoglycan (PG) synthesis through its transpeptidase (TP) domain, either with the peptidoglycan elongation machinery (PGEM), or with the wall itself. PBP2 also contains transmembrane (TM) and nonpenicillin binding (nPB) domains, which may mediate interactions with other morphogenetic proteins.
Fig. 3.
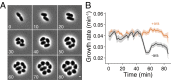
Growth rate is unaffected by substantial PBP2 depletion. (A) Time-lapse microscopy of E. coli TKL141 (∆mrdA Para-mrdA) cells growing during PBP2 depletion. Time is specified in minutes. Even though cells lose their rod-shaped morphology, they continue to grow at a rate comparable to wild-type cells until two to three divisions have taken place. (B) Growth rate during PBP2 depletion is quantitatively unaffected over more than two doubling times. Ara+ represents the nondepleted control, and Ara− represents cells undergoing PBP2 depletion. The shaded area represents SE. (Scale bar: 1 µm.)
Fig. 4.
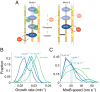
PBP2 activity is required throughout MreB-directed peptidoglycan synthesis. (A) Potential models for the role of PBP2 activity during cell wall synthesis. (Left) PBP2 is required for initiating strand synthesis by the rest of the cell wall synthesis machinery, but is not required for further synthesis-dependent MreB motion. (Right) Diffusing PBP2 molecules rapidly associate and disassociate with MreB-associated proteins to drive the progress of strand incorporation and MreB motion. (B) Cell growth rate decreases under mecillinam treatment in a concentration-dependent manner. Exponential phase cells (MG1655) were placed on agarose pads containing the indicated concentration of mecillinam and imaged for 15 min. To calculate growth rate, cell contours were extracted and cell length over time was fit to an exponential function. (C) MreB speed in TKL039 cells also decreases under mecillinam treatment in a concentration-dependent manner, indicating a requirement for active PBP2 molecules during the insertion of glycan strands.
Comment in
- Dynamic protein complexes for cell growth.
Banzhaf M, Typas A. Banzhaf M, et al. Proc Natl Acad Sci U S A. 2014 Mar 25;111(12):4355-6. doi: 10.1073/pnas.1402016111. Epub 2014 Mar 17. Proc Natl Acad Sci U S A. 2014. PMID: 24639518 Free PMC article. No abstract available.
Similar articles
- A central role for PBP2 in the activation of peptidoglycan polymerization by the bacterial cell elongation machinery.
Rohs PDA, Buss J, Sim SI, Squyres GR, Srisuknimit V, Smith M, Cho H, Sjodt M, Kruse AC, Garner EC, Walker S, Kahne DE, Bernhardt TG. Rohs PDA, et al. PLoS Genet. 2018 Oct 18;14(10):e1007726. doi: 10.1371/journal.pgen.1007726. eCollection 2018 Oct. PLoS Genet. 2018. PMID: 30335755 Free PMC article. - The transpeptidase PBP2 governs initial localization and activity of the major cell-wall synthesis machinery in E. coli.
Özbaykal G, Wollrab E, Simon F, Vigouroux A, Cordier B, Aristov A, Chaze T, Matondo M, van Teeffelen S. Özbaykal G, et al. Elife. 2020 Feb 20;9:e50629. doi: 10.7554/eLife.50629. Elife. 2020. PMID: 32077853 Free PMC article. - Principles of bacterial cell-size determination revealed by cell-wall synthesis perturbations.
Tropini C, Lee TK, Hsin J, Desmarais SM, Ursell T, Monds RD, Huang KC. Tropini C, et al. Cell Rep. 2014 Nov 20;9(4):1520-7. doi: 10.1016/j.celrep.2014.10.027. Epub 2014 Nov 6. Cell Rep. 2014. PMID: 25456140 Free PMC article. - Cell shape determination in Escherichia coli.
Osborn MJ, Rothfield L. Osborn MJ, et al. Curr Opin Microbiol. 2007 Dec;10(6):606-10. doi: 10.1016/j.mib.2007.09.004. Epub 2007 Nov 5. Curr Opin Microbiol. 2007. PMID: 17981077 Review. - MreB: pilot or passenger of cell wall synthesis?
White CL, Gober JW. White CL, et al. Trends Microbiol. 2012 Feb;20(2):74-9. doi: 10.1016/j.tim.2011.11.004. Epub 2011 Dec 7. Trends Microbiol. 2012. PMID: 22154164 Review.
Cited by
- Systematic perturbation of cytoskeletal function reveals a linear scaling relationship between cell geometry and fitness.
Monds RD, Lee TK, Colavin A, Ursell T, Quan S, Cooper TF, Huang KC. Monds RD, et al. Cell Rep. 2014 Nov 20;9(4):1528-37. doi: 10.1016/j.celrep.2014.10.040. Epub 2014 Nov 13. Cell Rep. 2014. PMID: 25456141 Free PMC article. - Response of Escherichia coli growth rate to osmotic shock.
Rojas E, Theriot JA, Huang KC. Rojas E, et al. Proc Natl Acad Sci U S A. 2014 May 27;111(21):7807-12. doi: 10.1073/pnas.1402591111. Epub 2014 May 12. Proc Natl Acad Sci U S A. 2014. PMID: 24821776 Free PMC article. - Mechanical strain sensing implicated in cell shape recovery in Escherichia coli.
Wong F, Renner LD, Özbaykal G, Paulose J, Weibel DB, van Teeffelen S, Amir A. Wong F, et al. Nat Microbiol. 2017 Jul 24;2:17115. doi: 10.1038/nmicrobiol.2017.115. Nat Microbiol. 2017. PMID: 28737752 Free PMC article. - Bacterial morphogenesis and the enigmatic MreB helix.
Errington J. Errington J. Nat Rev Microbiol. 2015 Apr;13(4):241-8. doi: 10.1038/nrmicro3398. Epub 2015 Jan 12. Nat Rev Microbiol. 2015. PMID: 25578957 - Dynamic protein complexes for cell growth.
Banzhaf M, Typas A. Banzhaf M, et al. Proc Natl Acad Sci U S A. 2014 Mar 25;111(12):4355-6. doi: 10.1073/pnas.1402016111. Epub 2014 Mar 17. Proc Natl Acad Sci U S A. 2014. PMID: 24639518 Free PMC article. No abstract available.
References
- Sauvage E, Kerff F, Terrak M, Ayala JA, Charlier P. The penicillin-binding proteins: Structure and role in peptidoglycan biosynthesis. FEMS Microbiol Rev. 2008;32(2):234–258. - PubMed
- Domínguez-Escobar J, et al. Processive movement of MreB-associated cell wall biosynthetic complexes in bacteria. Science. 2011;333(6039):225–228. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 1F32GM100677-01A1/GM/NIGMS NIH HHS/United States
- F32 GM100677/GM/NIGMS NIH HHS/United States
- P50 GM107615/GM/NIGMS NIH HHS/United States
- DP2OD006466/OD/NIH HHS/United States
- DP2 OD004389/OD/NIH HHS/United States
- R01 GM107384/GM/NIGMS NIH HHS/United States
- DP2OD004389/OD/NIH HHS/United States
- DP2 OD006466/OD/NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases