CRISPR-based technologies: prokaryotic defense weapons repurposed - PubMed (original) (raw)
Review
CRISPR-based technologies: prokaryotic defense weapons repurposed
Rebecca M Terns et al. Trends Genet. 2014 Mar.
Abstract
To combat potentially deadly viral infections, prokaryotic microbes enlist small RNA-based adaptive immune systems (CRISPR-Cas systems) that protect through sequence-specific recognition and targeted destruction of viral nucleic acids (either DNA or RNA depending on the system). Here, we summarize rapid progress made in redirecting the nuclease activities of these microbial immune systems to bind and cleave DNA or RNA targets of choice, by reprogramming the small guide RNAs of the various CRISPR-Cas complexes. These studies have demonstrated the potential of Type II CRISPR-Cas systems both as efficient and versatile genome-editing tools and as potent and specific regulators of gene expression in a broad range of cell types (including human) and organisms. Progress is also being made in developing a Type III RNA-targeting CRISPR-Cas system as a novel gene knockdown platform to investigate gene function and modulate gene expression for metabolic engineering in microbes.
Keywords: CRISPR; Cas; RNA silencing; biotechnology; genome editing; metabolic engineering.
Copyright © 2014 Elsevier Ltd. All rights reserved.
Figures
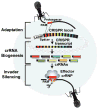
Figure 1
Overview of the CRISPR-Cas pathways. To obtain the ability to recognize and silence a specific virus or other foreign genetic element, a short fragment of invader DNA is acquired and integrated into the host CRISPR locus near the leader (Adaptation). The CRISPR locus contains copies of a short direct repeat sequence (black) that separate the invader-derived sequences (multiple colors). Cas genes encoding the protein components of the system are typically encoded adjacent to CRISPR loci (not shown). Protospacer adjacent motifs (PAMs) are found immediately adjacent to viral sequences selected for CRISPR integration (protospacers). CRISPR locus transcripts are processed to generate multiple individual crRNAs (crRNA Biogenesis). Mature crRNAs retain repeat sequence at the 5′ end (shown) and/or 3′ end of the invader-derived guide sequence (colored region). crRNAs associate with 1 to 6 Cas proteins (depending on the system) to form crRNA-Cas protein (crRNP) effector complexes. The crRNP specifically recognizes foreign DNA or RNA (depending on the system) via base-pairing of the crRNA and cleaves (Invader Silencing). PAMs provide important auxiliary signals for the recognition and silencing of invaders for some Type I and II DNA-targeting systems.
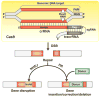
Figure 2
RNA-programmable genome editing by Type II Cas9 CRISPR-Cas systems. The Cas9 endonuclease (yellow) has two nuclease active sites (RuvC and HNH) that cleave opposing strands of a DNA molecule to generate a blunt-ended, double-strand DNA break (DSB). The cleavage is directed by the associated crRNA that contains a guide region that recognizes one strand of the target sequence by base-pairing. The enzyme requires a short
p
rotospacer
a
djacent
m
otif (PAM) immediately adjacent to the region of homology as an auxiliary target identification signal. Cas9 activity also requires a second RNA (tracrRNA) that base-pairs with the repeat-derived tag of the crRNA. The crRNA and tracrRNA can be combined through covalent linkage into a single-guide RNA (sgRNA). In human and many other cell types, the Cas9-induced, double-strand DNA breaks undergo repair by either non-homologous end joining (NHEJ) or homologous recombination (HR). NHEJ repair results in indels (short insertions or deletions) and provides a means to selectively disrupt or inactivate target genes. When provided with a homologous donor DNA, HR repair can affect sequence replacement to insert, modify or delete sequence at the site.
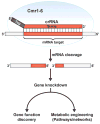
Figure 3
RNA-programmable gene knockdown by Type III-B Cmr CRISPR-Cas systems. The Cmr effector complex (blue) consists of six Cmr proteins and a crRNA. The guide region of the crRNA base-pairs with the homologus sequence in the mRNA target and the target RNA is cleaved by the complex. Applications of the Cmr system include RNA-directed gene knockdown to investigate gene function or to facilitate metabolic engineering.
Similar articles
- Characterization and Repurposing of Type I and Type II CRISPR-Cas Systems in Bacteria.
Hidalgo-Cantabrana C, Goh YJ, Barrangou R. Hidalgo-Cantabrana C, et al. J Mol Biol. 2019 Jan 4;431(1):21-33. doi: 10.1016/j.jmb.2018.09.013. Epub 2018 Sep 24. J Mol Biol. 2019. PMID: 30261168 Review. - RNA-guided genome editing à la carte.
Horvath P, Barrangou R. Horvath P, et al. Cell Res. 2013 Jun;23(6):733-4. doi: 10.1038/cr.2013.39. Epub 2013 Mar 12. Cell Res. 2013. PMID: 23478298 Free PMC article. - Genome editing using CRISPR, CAST, and Fanzor systems.
Song B, Bae S. Song B, et al. Mol Cells. 2024 Jul;47(7):100086. doi: 10.1016/j.mocell.2024.100086. Epub 2024 Jun 21. Mol Cells. 2024. PMID: 38909984 Free PMC article. Review. - Use of CRISPR/Cas Genome Editing Technology for Targeted Mutagenesis in Rice.
Xu R, Wei P, Yang J. Xu R, et al. Methods Mol Biol. 2017;1498:33-40. doi: 10.1007/978-1-4939-6472-7_3. Methods Mol Biol. 2017. PMID: 27709567 - [An history of the CRISPR-Cas systems discovery].
Pourcel C. Pourcel C. Biol Aujourdhui. 2017;211(4):247-254. doi: 10.1051/jbio/2018001. Epub 2018 Jun 29. Biol Aujourdhui. 2017. PMID: 29956651 French.
Cited by
- Type III-A CRISPR systems as a versatile gene knockdown technology.
Woodside WT, Vantsev N, Catchpole RJ, Garrett SC, Olson S, Graveley BR, Terns MP. Woodside WT, et al. RNA. 2022 Aug;28(8):1074-1088. doi: 10.1261/rna.079206.122. Epub 2022 May 26. RNA. 2022. PMID: 35618430 Free PMC article. - RNAi screening comes of age: improved techniques and complementary approaches.
Mohr SE, Smith JA, Shamu CE, Neumüller RA, Perrimon N. Mohr SE, et al. Nat Rev Mol Cell Biol. 2014 Sep;15(9):591-600. doi: 10.1038/nrm3860. Nat Rev Mol Cell Biol. 2014. PMID: 25145850 Free PMC article. Review. - Cas9-based genome editing in Xenopus tropicalis.
Nakayama T, Blitz IL, Fish MB, Odeleye AO, Manohar S, Cho KW, Grainger RM. Nakayama T, et al. Methods Enzymol. 2014;546:355-75. doi: 10.1016/B978-0-12-801185-0.00017-9. Methods Enzymol. 2014. PMID: 25398349 Free PMC article. - The history and market impact of CRISPR RNA-guided nucleases.
van Erp PB, Bloomer G, Wilkinson R, Wiedenheft B. van Erp PB, et al. Curr Opin Virol. 2015 Jun;12:85-90. doi: 10.1016/j.coviro.2015.03.011. Epub 2015 Apr 26. Curr Opin Virol. 2015. PMID: 25914022 Free PMC article. - Programmable type III-A CRISPR-Cas DNA targeting modules.
Ichikawa HT, Cooper JC, Lo L, Potter J, Terns RM, Terns MP. Ichikawa HT, et al. PLoS One. 2017 Apr 25;12(4):e0176221. doi: 10.1371/journal.pone.0176221. eCollection 2017. PLoS One. 2017. PMID: 28441427 Free PMC article.
References
- Labrie SJ, et al. Bacteriophage resistance mechanisms. Nature reviews Microbiology. 2010;8:317–327. - PubMed
- Westra ER, et al. The CRISPRs, they are a-changin’: how prokaryotes generate adaptive immunity. Annual review of genetics. 2012;46:311–339. - PubMed
- Samson JE, et al. Revenge of the phages: defeating bacterial defences. Nature reviews Microbiology. 2013;11:675–687. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM054682/GM/NIGMS NIH HHS/United States
- R01 GM099876/GM/NIGMS NIH HHS/United States
- R01 GM54682/GM/NIGMS NIH HHS/United States
- R01 GM99876/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources