Role of molecular charge in nucleocytoplasmic transport - PubMed (original) (raw)
Role of molecular charge in nucleocytoplasmic transport
Alexander Goryaynov et al. PLoS One. 2014.
Abstract
Transport of genetic materials and proteins between the nucleus and cytoplasm of eukaryotic cells is mediated by nuclear pore complexes (NPCs). A selective barrier formed by phenylalanine-glycine (FG) nucleoporins (Nups) with net positive charges in the NPC allows for passive diffusion of signal-independent small molecules and transport-receptor facilitated translocation of signal-dependent cargo molecules. Recently, negative surface charge was postulated to be another essential criterion for selective passage through the NPC. However, the charge-driven mechanism in determining the transport kinetics and spatial transport route for either passive diffusion or facilitated translocation remains obscure. Here we employed high-speed single-molecule fluorescence microscopy with an unprecedented spatiotemporal resolution of 9 nm and 400 µs to uncover these mechanistic fundamentals for nuclear transport of charged substrates through native NPCs. We found that electrostatic interaction between negative surface charges on transiting molecules and the positively charged FG Nups, although enhancing their probability of binding to the NPC, never plays a dominant role in determining their nuclear transport mode or spatial transport route. A 3D reconstruction of transport routes revealed that small signal-dependent endogenous cargo protein constructs with high positive surface charges that are destined to the nucleus, rather than repelled from the NPC as suggested in previous models, passively diffused through an axial central channel of the NPC in the absence of transport receptors. Finally, we postulated a comprehensive map of interactions between transiting molecules and FG Nups during nucleocytoplasmic transport by combining the effects of molecular size, signal and surface charge.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
Figure 1. Transport of single molecules through a single NPC tracked by SPEED microscopy and further characterized with 2D to 3D deconvolution algorithms.
(A) Diagram of the illumination of a single GFP-NPC (blue) and the tracking of single fluorescent in-transit molecules (red dots) through this NPC in cells by SPEED microscopy. SPEED microscopy generates an inclined illumination point spread function (iPSF forms an angle of 45° to the z direction) in the focal plane (between the double light blue lines) at the equatorial plane of a cell nucleus. C, cytoplasm. N, nucleus. (B) A well-isolated fluorescent spot of a single GFP-NPC (green) overlaid with a single in-transit fluorescent molecule (SM, red spot, here using Alexa Fluor 647 labeled single GFP molecule as an example) captured by SPEED microscopy. A typical nuclear transport event of a single molecule (red spot) from the nucleus to the cytoplasm is captured at 0.4 ms per frame. Numbers denote time in milliseconds. Scale bar, 1 µm. (C) Using a 2D Gaussian fit of isolated fluorescent spots, the trajectories of an in-transit molecule (red dots) and the centroid of the NPC (the blue dot) were obtained and then overlaid onto the NPC architecture (blue). Clearly, the transiting molecule interacted with the NPC from 0.8 ms to 2.8 ms. N, the nucleoplasmic side of the NPC. C, the cytoplasmic side of the NPC. (D) The diagram illustrates the 2D to 3D deconvolution process. In principle, 2D spatial locations (x, y) projected from 3D locations (x, y, z) in a Cartesian coordinate system retain all of the information about the 3D locations that can also be encoded in a cylindrical coordinate system (R, θ, x). Following an established approach, here a deconvolution process (or a back-projection process) was set up between a simplified cylindrical coordinate system (R, constant, x) and the obtained 2D spatial locations (x, y). Detailed deconvolution process can be found in previous publications , .
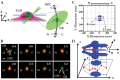
Figure 2. 3D passive diffusion routes of differently charged GFPs.
(A) −30 GFP. Calculated electrostatic surface potentials of GFP variants, ranging from −15 kT/e (dark blue) to +15 kT/e (dark red); neutral charge 0 is shown in green (I). Superimposed plots of thousands of localizations of single molecules located primarily within a rectangular area of 240×160 nm (II). N, the nucleoplasmic side of the NPC. C, the cytoplasmic side of the NPC. The locations in each 10×10 nm area were quantized and filtered with a Gaussian blur function to generate the 2D probability density map overlaid onto the NPC architecture (light blue). The highest density was 1×105 locations/µm2, and the lowest was 0 locations/µm2, shown in gray (III). A 3D probability density map generated using 2D to 3D deconvolution algorithms (red cloud; brighter color indicates higher density) is shown in both side-view and a top-view orientations superimposed on the NPC architecture (blue). The length of the path and the diameter at the central plane of the NPC were measured and marked in nanometers (IV). N, the nucleoplasmic side of the NPC. C, the cytoplasmic side of the NPC. (B–C) Nucleocytoplasmic transport pathways of −7 GFP and +36 GFP.
Figure 3. 3D pathways of Imp β1 alone and Imp α/Imp β1 in complex with differently charged GFP cargo (−30 NLS-GFP), −7 NLS-GFP and +36 NLS-GFP).
(A) Imp β1 alone. Calculated electrostatic surface potentials of Imp β1 range from −15 kT/e (dark blue) to +15 kT/e (dark red), and neutral charge 0 is shown in green (I). Superimposed plots of localizations of single molecules located primarily within a rectangular area of 240×160 nm (II). N, the nucleoplasmic side of the NPC; C, the cytoplasmic side of the NPC. The locations in each 10×10 nm area were quantized and filtered with a Gaussian blur function to generate the 2D probability density map overlaid onto the NPC architecture (light blue). The highest density was 1.7×105 locations/µm2 and the lowest was 0 locations/µm2, shown in gray (III). A 3D probability density map (green cloud; brighter color indicates higher density) is shown in both side-view and a top-view orientations superimposed on the NPC architecture (blue). The length of pathway and the diameter at the central plane of NPC was measured and is labeled in nanometers. N, the nucleoplasmic side of the NPC. C, the cytoplasmic side of the NPC. (B–D) Nucleocytoplasmic transport pathways of −30NLS-GFP/Impα/Impβ1, −7NLS-GFP/Impα/Impβ1 and +36NLS-GFP/Impα/Impβ1.
Figure 4. 3D transport pathways of a positively charged ribosomal protein rpL23 through the NPC with and without Imp β1.
(A) 3D transport routes of rpL23 alone. N, the nucleoplasmic side of the NPC. C, the cytoplasmic side of the NPC. (B) 3D transport routes of the cargo complex of rpL23/Imp β1. (C) Time-courses of net nuclear accumulations of rpL23 alone and rpL23/Imp β1. The graphs depict average intensity increase in the nuclei as the permeabilized cells were incubated with rpL23 (red open dots) or rpL23/Imp β1 (black open dots). The time-course data were fitted to an exponential function of the form f(t) = f_max(1−_e_−_kt), where t is time, f(t) is the nuclear fluorescence, fmax is the endpoint of the reaction and k is the first-order rate constant. The fits revealed that the initial transport rate (r = kfmax) of rpL23/Imp β1 was approximately three-fold higher than that of rpL23 alone.
Figure 5. Summary of the charge effect on nucleocytoplasmic transport.
(A) Charge effect on passive diffusion. Small positively charged molecules (<40 kDa, red dots) permeate the NPC through the central channel just as small negatively charged (blue dots), but with a shorter and narrower route. (B) Charge effect on facilitated transport. Regardless of the cargo’s charge, transiting cargo complexes (transport receptor/cargo) follow the pathways of the transport receptor through the FG filamentous regions at the periphery around the central channel.
Similar articles
- The selective permeability barrier in the nuclear pore complex.
Li C, Goryaynov A, Yang W. Li C, et al. Nucleus. 2016 Sep 2;7(5):430-446. doi: 10.1080/19491034.2016.1238997. Epub 2016 Sep 27. Nucleus. 2016. PMID: 27673359 Free PMC article. Review. - Distinct, but not completely separate spatial transport routes in the nuclear pore complex.
Yang W. Yang W. Nucleus. 2013 May-Jun;4(3):166-75. doi: 10.4161/nucl.24874. Epub 2013 May 1. Nucleus. 2013. PMID: 23669120 Free PMC article. Review. - Single-molecule studies of nucleocytoplasmic transport: from one dimension to three dimensions.
Goryaynov A, Ma J, Yang W. Goryaynov A, et al. Integr Biol (Camb). 2012 Jan;4(1):10-21. doi: 10.1039/c1ib00041a. Epub 2011 Oct 24. Integr Biol (Camb). 2012. PMID: 22020388 Free PMC article. Review. - SPEED Microscopy and Its Application in Nucleocytoplasmic Transport.
Ma J, Kelich JM, Yang W. Ma J, et al. Methods Mol Biol. 2016;1411:503-18. doi: 10.1007/978-1-4939-3530-7_31. Methods Mol Biol. 2016. PMID: 27147062 Free PMC article. - 'Natively unfolded' nucleoporins in nucleocytoplasmic transport: clustered or evenly distributed?
Yang W. Yang W. Nucleus. 2011 Jan-Feb;2(1):10-6. doi: 10.4161/nucl.2.1.13818. Nucleus. 2011. PMID: 21647294 Free PMC article.
Cited by
- Physics of the Nuclear Pore Complex: Theory, Modeling and Experiment.
Hoogenboom BW, Hough LE, Lemke EA, Lim RYH, Onck PR, Zilman A. Hoogenboom BW, et al. Phys Rep. 2021 Jul 25;921:1-53. doi: 10.1016/j.physrep.2021.03.003. Epub 2021 Mar 24. Phys Rep. 2021. PMID: 35892075 Free PMC article. - Particle transport through hydrogels is charge asymmetric.
Zhang X, Hansing J, Netz RR, DeRouchey JE. Zhang X, et al. Biophys J. 2015 Feb 3;108(3):530-9. doi: 10.1016/j.bpj.2014.12.009. Biophys J. 2015. PMID: 25650921 Free PMC article. - Reply to 'Deconstructing transport-distribution reconstruction in the nuclear-pore complex'.
Ruba A, Kelich J, Ma J, Yang W. Ruba A, et al. Nat Struct Mol Biol. 2018 Dec;25(12):1062-1064. doi: 10.1038/s41594-018-0162-1. Epub 2018 Dec 5. Nat Struct Mol Biol. 2018. PMID: 30518846 Free PMC article. No abstract available. - Genome-Wide Identification, Expression Profile and Evolution Analysis of Karyopherin β Gene Family in Solanum tuberosum Group Phureja DM1-3 Reveals Its Roles in Abiotic Stresses.
Xu Y, Liu L, Zhao P, Tong J, Zhong N, Zhang H, Liu N. Xu Y, et al. Int J Mol Sci. 2020 Jan 31;21(3):931. doi: 10.3390/ijms21030931. Int J Mol Sci. 2020. PMID: 32023817 Free PMC article. - High-resolution imaging reveals new features of nuclear export of mRNA through the nuclear pore complexes.
Kelich JM, Yang W. Kelich JM, et al. Int J Mol Sci. 2014 Aug 20;15(8):14492-504. doi: 10.3390/ijms150814492. Int J Mol Sci. 2014. PMID: 25141104 Free PMC article. Review.
References
- Powers M, Forbes D (2012) Nuclear transport: beginning to gel? Curr Biol 22: R1006–1009. - PubMed
- Görlich D, Kutay U (1999) Transport between the cell nucleus and the cytoplasm. Annu Rev Cell Dev Biol 15: 607–660. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources