New routes to targeted therapy of intrahepatic cholangiocarcinomas revealed by next-generation sequencing - PubMed (original) (raw)
doi: 10.1634/theoncologist.2013-0352. Epub 2014 Feb 21.
Kai Wang, Laurie Gay, Rami Al-Rohil, Janne V Rand, David M Jones, Hwa J Lee, Christine E Sheehan, Geoff A Otto, Gary Palmer, Roman Yelensky, Doron Lipson, Deborah Morosini, Matthew Hawryluk, Daniel V T Catenacci, Vincent A Miller, Chaitanya Churi, Siraj Ali, Philip J Stephens
Affiliations
- PMID: 24563076
- PMCID: PMC3958461
- DOI: 10.1634/theoncologist.2013-0352
New routes to targeted therapy of intrahepatic cholangiocarcinomas revealed by next-generation sequencing
Jeffrey S Ross et al. Oncologist. 2014 Mar.
Abstract
Background: Intrahepatic cholangiocarcinoma (ICC) is a subtype of primary liver cancer that is rarely curable by surgery and is rapidly increasing in incidence. Relapsed ICC has a poor prognosis, and current systemic nontargeted therapies are commonly extrapolated from those used in other gastrointestinal malignancies. We hypothesized that genomic profiling of clinical ICC samples would identify genomic alterations that are linked to targeted therapies and that could facilitate a personalized approach to therapy.
Methods: DNA sequencing of hybridization-captured libraries was performed for 3,320 exons of 182 cancer-related genes and 36 introns of 14 genes frequently rearranged in cancer. Sample DNA was isolated from 40 μm of 28 formalin-fixed paraffin-embedded ICC specimens and sequenced to high coverage.
Results: The most commonly observed alterations were within ARID1A (36%), IDH1/2 (36%), and TP53 (36%) as well as amplification of MCL1 (21%). Twenty cases (71%) harbored at least one potentially actionable alteration, including FGFR2 (14%), KRAS (11%), PTEN (11%), CDKN2A (7%), CDK6 (7%), ERBB3 (7%), MET (7%), NRAS (7%), BRCA1 (4%), BRCA2 (4%), NF1 (4%), PIK3CA (4%), PTCH1 (4%), and TSC1 (4%). Four (14%) of the ICC cases featured novel gene fusions involving the tyrosine kinases FGFR2 and NTRK1 (FGFR2-KIAA1598, FGFR2-BICC1, FGFR2-TACC3, and RABGAP1L-NTRK1).
Conclusion: Two thirds of patients in this study harbored genomic alterations that are associated with targeted therapies and that have the potential to personalize therapy selection for to individual patients.
Keywords: Driver mutations; Intrahepatic cholangiocarcinoma; Next-generation sequencing; Targeted therapy.
Conflict of interest statement
Disclosures of potential conflicts of interest may be found at the end of this article.
Figures
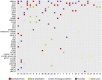
Figure 1.
Tile plot of genomic alterations in 28 cases of intrahepatic cholangiocarcinoma.
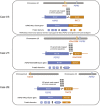
Figure 2.
Diagram of FGFR2 gene fusions in three cases of intrahepatic cholangiocarcinoma.
Figure 3.
Histology and list of genomic alterations in three cases of intrahepatic cholangiocarcinoma featuring FGFR2 gene fusions detected by next-generation sequencing.
References
- Vasilieva LE, Papadhimitriou SI, Dourakis SP. Modern diagnostic approaches to cholangiocarcinoma. Hepatobiliary Pancreat Dis Int. 2012;11:349–359. - PubMed
- Sempoux C, Jibara G, Ward SC, et al. Intrahepatic cholangiocarcinoma: New insights in pathology. Semin Liver Dis. 2011;31:49–60. - PubMed
- Poultsides GA, Zhu AX, Choti MA, et al. Intrahepatic cholangiocarcinoma. Surg Clin North Am. 2010;90:817–837. - PubMed
- Bartlett DL. Intrahepatic cholangiocarcinoma: A worthy challenge. Cancer J. 2009;15:255–256. - PubMed
- Kobayashi M, Ikeda K, Saitoh S, et al. Incidence of primary cholangiocellular carcinoma of the liver in japanese patients with hepatitis C virus-related cirrhosis. Cancer. 2000;88:2471–2477. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous