Comprehensive analysis of the specificity of transcription activator-like effector nucleases - PubMed (original) (raw)
. 2014 Apr;42(8):5390-402.
doi: 10.1093/nar/gku155. Epub 2014 Feb 24.
Gwendoline Dubois, Julien Valton, Séverine Thomas, Stefano Stella, Alan Maréchal, Stéphanie Langevin, Nassima Benomari, Claudia Bertonati, George H Silva, Fayza Daboussi, Jean-Charles Epinat, Guillermo Montoya, Aymeric Duclert, Philippe Duchateau
Affiliations
- PMID: 24569350
- PMCID: PMC4005648
- DOI: 10.1093/nar/gku155
Comprehensive analysis of the specificity of transcription activator-like effector nucleases
Alexandre Juillerat et al. Nucleic Acids Res. 2014 Apr.
Abstract
A key issue when designing and using DNA-targeting nucleases is specificity. Ideally, an optimal DNA-targeting tool has only one recognition site within a genomic sequence. In practice, however, almost all designer nucleases available today can accommodate one to several mutations within their target site. The ability to predict the specificity of targeting is thus highly desirable. Here, we describe the first comprehensive experimental study focused on the specificity of the four commonly used repeat variable diresidues (RVDs; NI:A, HD:C, NN:G and NG:T) incorporated in transcription activator-like effector nucleases (TALEN). The analysis of >15 500 unique TALEN/DNA cleavage profiles allowed us to monitor the specificity gradient of the RVDs along a TALEN/DNA binding array and to present a specificity scoring matrix for RVD/nucleotide association. Furthermore, we report that TALEN can only accommodate a relatively small number of position-dependent mismatches while maintaining a detectable activity at endogenous loci in vivo, demonstrating the high specificity of these molecular tools. We thus envision that the results we provide will allow for more deliberate choices of DNA binding arrays and/or DNA targets, extending our engineering capabilities.
Figures
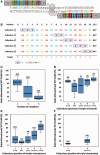
Figure 1.
Gene disruption-relative activities of collection of TALEN at the integrated GFP locus. (A) Schematic representation of the WT GFP TALEN on the chromosomal target. (B) Collections of TALE used at the chromosomal GFP locus derived by mutation of the right DNA binding domain. X represents any of the four, namely, NI, HD, NN and NG RVD. Positions are numbered relative to the first thymine of the target (T0). (C) Influence of the number of mismatches on the GFP disruption. Activity ratio between the mismatched and the WT TALEN is represented on a boxplot, indicating the median (thick bar), quartiles (box) and extreme values (r = −0.68, P = 4e−16). Mismatches are defined relative to the NI:A, HD:C, NN:G and NG:T codes. The size of the sample is indicated in brackets. (D) Boxplot representation, including the median (thick bar), quartiles (box) and extreme values, of the activity ratio between the mismatched and the WT TALEN in function of the collections for one mismatch. P = 0.08 (Kruskal–Wallis test). (E) Same as for (D) but for two mismatches. P = 4e−4. (F) Same as in (D) but for three mismatches. P = 2e−4.
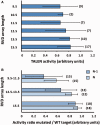
Figure 2.
Design of experimental setup and optimal TALEN array length for the nuclease activity screening. (A) Activities of 52 TALEN pairs measured in vivo, in function of the repeat array length. Error bars represent standard errors. (B) Activities of 15 TALEN pairs on targets randomized in their positions N and N-1. Error bars represent standard errors. An analysis of variance demonstrates a significant effect of the TALEN length on specificity, position N: r = 0.68, P = 4e−6 (Kruskal–Wallis test); position N-1: r = 0.7, P = 7e−6). The yeast activity assay is based on the single-strand annealing (SSA) pathway used after the creation of a DSB by the TALEN in the target sequence. Target sites were designed to allow TALEN use in the homodimer format. All TALEN pairs showed a significant activity. The number of TALEN/target pairs for each class is indicated in brackets.
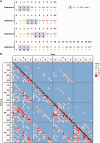
Figure 3.
Setup of collections used in the large-scale experiments and graphical representation of activity results from Collection 6. (A) Collections of TALE and targets used for the study in yeast, where X represents any of the four NI, HD, NN and NG RVD and N any of the four A, C, G and T bases. Collections 6–8 are composed of arrays containing 9.5 repeats and Collection 9 is composed of arrays of 18.5. The TALEN collections were used in the homodimer format. (B) Heatmap showing the activity of the 64 TALEN of Collection 6 on the 64 corresponding targets. The outer line of text of the target (abscissa) represents the first nucleotide of the NNN triplet, the middle line of text represents the second nucleotide and the innermost line of text represents the third nucleotide. Likewise for the RVD array (XXX) on the ordinate. Red corresponds to maximum activity, whereas blue corresponds to no activity. The diagonal with framed squares represents the NI:A, HD:C, NN:G and NG:T pairings. In the case of a perfect one to one RVD/nucleotide association code, activity should be recovered on the diagonal only.
Figure 4.
Graphical representation of RVD specificities. (A) Specificity measured for positions 1–7. The level of gray (black = 1, white = 0) represents the relative activity compared with the HD:C, NG:T NI:A and NN:G RVD/nucleotide pairing code. (B) Average specificity of the four HD, NG, NI and NN RVDs on the first 7 positions. (C) Logo representation of the global specificity matrix. Logo was generated using WebLogo (
http://weblogo.berkeley.edu/logo.cgi
). Values for relative specificities are presented in
Supplementary Table S4
.
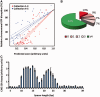
Figure 5.
Effect of target mutations on TALEN activity in mammalian cells. (A) Correlation between experimental relative activities represented by the percentage of GFP-negative cells in the mammalian gene-targeting assay and scoring using the matrix presented in Figure 4B (Collections 1, 2 and 3 are represented in red: r = 0.81, P = 4e−16, and Collections 4 and 5 are represented in blue: r = 0.84, P = 3e−12). Linear regressions are presented for both subsets and 95% confidence intervals are represented by dashed lines. (B) Pie chart representation of the percentage of TALEN composed of 15.5 RVDs that will have, in the human genome, potential off-site targets containing no, one, two, three or four and more mismatches when considering a test set of 15 000 putative TALEN. All possible combinations of half TALEN (left + right, left + left, right + right) with a spacer length ranging from 9 to 30 bp were taken into account. (C) A collection of 33 targets comprising two AvrBs3 target sequences facing each other on both DNA strand with spacer length (between the two targets) ranging from 5 to 40 bp were designed and assayed in the CHO-K1 SSA assay to determine the optimal cleavage conditions. Targets containing a spacer of 21 and 35 bp were absent from the study.
Similar articles
- Targeting G with TAL effectors: a comparison of activities of TALENs constructed with NN and NK repeat variable di-residues.
Christian ML, Demorest ZL, Starker CG, Osborn MJ, Nyquist MD, Zhang Y, Carlson DF, Bradley P, Bogdanove AJ, Voytas DF. Christian ML, et al. PLoS One. 2012;7(9):e45383. doi: 10.1371/journal.pone.0045383. Epub 2012 Sep 24. PLoS One. 2012. PMID: 23028976 Free PMC article. - Optimized tuning of TALEN specificity using non-conventional RVDs.
Juillerat A, Pessereau C, Dubois G, Guyot V, Maréchal A, Valton J, Daboussi F, Poirot L, Duclert A, Duchateau P. Juillerat A, et al. Sci Rep. 2015 Jan 30;5:8150. doi: 10.1038/srep08150. Sci Rep. 2015. PMID: 25632877 Free PMC article. - SAPTA: a new design tool for improving TALE nuclease activity.
Lin Y, Fine EJ, Zheng Z, Antico CJ, Voit RA, Porteus MH, Cradick TJ, Bao G. Lin Y, et al. Nucleic Acids Res. 2014 Apr;42(6):e47. doi: 10.1093/nar/gkt1363. Epub 2014 Jan 16. Nucleic Acids Res. 2014. PMID: 24442582 Free PMC article. - Transcription activator-like effector nucleases (TALENs): a highly efficient and versatile tool for genome editing.
Sun N, Zhao H. Sun N, et al. Biotechnol Bioeng. 2013 Jul;110(7):1811-21. doi: 10.1002/bit.24890. Epub 2013 Apr 7. Biotechnol Bioeng. 2013. PMID: 23508559 Review. - TALENs: a widely applicable technology for targeted genome editing.
Joung JK, Sander JD. Joung JK, et al. Nat Rev Mol Cell Biol. 2013 Jan;14(1):49-55. doi: 10.1038/nrm3486. Epub 2012 Nov 21. Nat Rev Mol Cell Biol. 2013. PMID: 23169466 Free PMC article. Review.
Cited by
- Efficient multitool/multiplex gene engineering with TALE-BE.
Boyne A, Yang M, Pulicani S, Feola M, Tkach D, Hong R, Duclert A, Duchateau P, Juillerat A. Boyne A, et al. Front Bioeng Biotechnol. 2022 Nov 10;10:1033669. doi: 10.3389/fbioe.2022.1033669. eCollection 2022. Front Bioeng Biotechnol. 2022. PMID: 36440442 Free PMC article. - Context influences on TALE-DNA binding revealed by quantitative profiling.
Rogers JM, Barrera LA, Reyon D, Sander JD, Kellis M, Joung JK, Bulyk ML. Rogers JM, et al. Nat Commun. 2015 Jun 11;6:7440. doi: 10.1038/ncomms8440. Nat Commun. 2015. PMID: 26067805 Free PMC article. - Description of CRISPR/Cas9 development and its prospect in hepatocellular carcinoma treatment.
Wu X, Ma W, Mei C, Chen X, Yao Y, Liu Y, Qin X, Yuan Y. Wu X, et al. J Exp Clin Cancer Res. 2020 Jun 1;39(1):97. doi: 10.1186/s13046-020-01603-0. J Exp Clin Cancer Res. 2020. PMID: 32487115 Free PMC article. Review. - Advances in Recombinant Adeno-Associated Virus Vectors for Neurodegenerative Diseases.
Li L, Vasan L, Kartono B, Clifford K, Attarpour A, Sharma R, Mandrozos M, Kim A, Zhao W, Belotserkovsky A, Verkuyl C, Schmitt-Ulms G. Li L, et al. Biomedicines. 2023 Oct 8;11(10):2725. doi: 10.3390/biomedicines11102725. Biomedicines. 2023. PMID: 37893099 Free PMC article. Review. - A CRISPR/Cas9 Toolbox for Multiplexed Plant Genome Editing and Transcriptional Regulation.
Lowder LG, Zhang D, Baltes NJ, Paul JW 3rd, Tang X, Zheng X, Voytas DF, Hsieh TF, Zhang Y, Qi Y. Lowder LG, et al. Plant Physiol. 2015 Oct;169(2):971-85. doi: 10.1104/pp.15.00636. Epub 2015 Aug 21. Plant Physiol. 2015. PMID: 26297141 Free PMC article.
References
- DeFrancesco L. Move over ZFNs. Nat. Biotechnol. 2011;29:681–684. - PubMed
- Bogdanove AJ, Voytas DF. TAL effectors: customizable proteins for DNA targeting. Science. 2011;333:1843–1846. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources