Atomic model of the F420-reducing [NiFe] hydrogenase by electron cryo-microscopy using a direct electron detector - PubMed (original) (raw)
Atomic model of the F420-reducing [NiFe] hydrogenase by electron cryo-microscopy using a direct electron detector
Matteo Allegretti et al. Elife. 2014.
Abstract
The introduction of direct electron detectors with higher detective quantum efficiency and fast read-out marks the beginning of a new era in electron cryo-microscopy. Using the FEI Falcon II direct electron detector in video mode, we have reconstructed a map at 3.36 Å resolution of the 1.2 MDa F420-reducing hydrogenase (Frh) from methanogenic archaea from only 320,000 asymmetric units. Videos frames were aligned by a combination of image and particle alignment procedures to overcome the effects of beam-induced motion. The reconstructed density map shows all secondary structure as well as clear side chain densities for most residues. The full coordination of all cofactors in the electron transfer chain (a [NiFe] center, four [4Fe4S] clusters and an FAD) is clearly visible along with a well-defined substrate access channel. From the rigidity of the complex we conclude that catalysis is diffusion-limited and does not depend on protein flexibility or conformational changes. DOI: http://dx.doi.org/10.7554/eLife.01963.001\.
Keywords: Methanothermobacter marburgensis; [NiFe] hydrogenase; cryo-electron microscopy; methanogenesis.
Conflict of interest statement
WK: Reviewing editor, eLife.
The other authors declare that no competing interests exist.
Figures
Figure 1.. Cryo-EM data collection of the Frh complex and CTF correction.
(A) A typical electron micrograph recorded with the Falcon II camera on the FEI Tecnai Polara operated at 300 kV. The defocus was determined by CTFFIND3 to be 0.9 µm (Mindell and Grigorieff, 2003). The particles are clearly visible and easy to box. Scale bar, 25 nm. (B) CTF of boxed particles. The inset shows a zoom of the high-resolution range. Thon rings are visible beyond 80% of the Nyquist frequency at 0.3 Å−1. DOI:
http://dx.doi.org/10.7554/eLife.01963.003
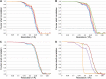
Figure 2.. Fourier shell correlation (FSC) curves for different refinement strategies.
All refinements were performed with the gold standard procedure in RELION (Scheres and Chen, 2012). A post-processing procedure (Chen et al., 2013) was applied unless otherwise indicated. The dotted line is at FSC 0.143, used to determine the resolution from comparing two independently refined half data sets (Rosenthal and Henderson, 2003). (A) Comparison of different alignment procedures. Blue, average of 20 unaligned frames. Purple, 20 frames aligned with the statistical video processing procedure (Bai et al., 2013). Red, refinement after aligning frames with the area-motion correction software (Li et al., 2013). Gold, combination of the latter two alignment procedures. (B) Effect of radiation damage. Blue, 20 frames (3.69 Å); purple, 16 frames (3.69 Å); red, 12 frames (3.60 Å); gold, 8 frames (3.43 Å); green, 6 frames (3.39 Å). All curves were obtained by the combination approach described above. (C) Data set quality. Particle images of sub-standard quality were omitted from the original data set of 33,590 images, yielding a smaller dataset of 26,635 particles (‘Results’). Refinement using 8 frames of the reduced data set (green, 3.39 Å). The improvement is clear in comparison with the full dataset (blue, 3.43 Å). A further improvement (red, 3.36 Å) resulted from using 6 instead of 8 frames. (D) Post-processing to determine the resolution and B-factor (Chen et al., 2013) of the final map from the 6-frame refinement. The raw unmasked map (purple) indicates a resolution of 3.52 Å; the map masked with a soft mask (red) indicates the final resolution of 3.36 Å. The gold curve shows the FSC for the two half data sets with randomized phases beyond 4.5 Å. Subtraction of the gold curve from the red curve yields the green curve, which indicates the true map resolution corrected for over-aggressive masking. The close correspondence of the red and green curves shows that the used soft mask did not introduce spurious correlation and the true map resolution is 3.36 Å. A B factor of −156 Å2 was determined and applied to sharpen the map. DOI:
http://dx.doi.org/10.7554/eLife.01963.004
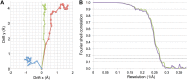
Figure 3.. Specimen movement as detected by recording images in video mode.
The motion correction software indicates a large movement at the beginning of the exposure. (A) Video frame alignment for three separate micrographs. Each spot represents one frame. The drift plots show that the movement between the first and second frame is considerably higher than in subsequent frames, although some micrographs (red and green) indicate much higher drift than others (blue). (B) FSC curves of a refinement with and without the first video frame. The violet curve represents a refinement of frames 1–17, the green curve frames 2–17. Although at FSC 0.143 the resolution is the same for for both maps (3.69 Å), the green curve without the first frame clearly shows a higher FSC in the whole resolution range. DOI:
http://dx.doi.org/10.7554/eLife.01963.005
Figure 4.. The 3.36 Å map with each of the 12 heterotrimers in a different colour.
DOI:
http://dx.doi.org/10.7554/eLife.01963.006
Figure 5.. Effect of radiation damage.
Helix 124–144 of FrhA in the map calculated from (A) video frames 1–6 (3.36 Å), (B) frames 8–13, pre-irradiated by ∼24 e/Å2 (3.94 Å), and (C) frames 15–20, pre-irradiated by ∼49 e/Å2 (4.16 Å). Note that side chain density is lacking for Asp125 and Glu132 already in the first map. DOI:
http://dx.doi.org/10.7554/eLife.01963.007
Figure 6.. EM map of Frh at 3.36 Å resolution.
(A) Slice through an FrhABG heterotrimer at the level of the electron transfer chain. (B) The same slice with atomic model. In this and other figures the carbons of FrhA are green, FrhG magenta, and FrhB blue. Green and orange spheres indicate the [NiFe] center in FrhA; the [4Fe4S] clusters in FrhG and FrhB are shown as orange and yellow spheres. The FAD in FrhB is shown as a stick model with yellow carbons. DOI:
http://dx.doi.org/10.7554/eLife.01963.008
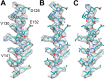
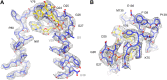
Figure 7.. Two helices of the 4-helix bundle in FrhA (Leu92-Ala114 and Val276-Glu300) without and with the model.
Note the absence of side chain density for glutamate and aspartate side chains. DOI:
http://dx.doi.org/10.7554/eLife.01963.010
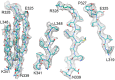
Figure 8.. Beta sheet 319-348 of FrhA.
(A) Top view, (B–D) side views of individual strands 341–348, 328–339 and 319–327, rotated by 90° relative to (A). DOI:
http://dx.doi.org/10.7554/eLife.01963.011
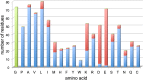
Figure 9.. Resolved amino acid side chains in the 3.36 Å map.
The histogram shows the number of residues on the y-axis and the amino acid (one letter code) on the x-axis. Blue bars indicate fully resolved side chains, red bars indicate side chains without or with ambiguous map density. Glycine residues (no side chain) are indicated in green for completeness. Negatively charged side chains of aspartate (D) and glutamate (E) are almost all missing. In contrast, side chains of hydrophobic residues like valine (V), leucine (L), isoleucine (I), phenylalanine (F), tyrosine (Y), and tryptophan (W), are nearly all visible. Excluding glycine, aspartate and glutamate, 86% of side chains are well visible. Unresolved side chains other than aspartate and glutamate are mostly located on the surface. DOI:
http://dx.doi.org/10.7554/eLife.01963.012
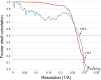
Figure 10.. Correlation between map and model.
The blue line shows the FSC between the final cryo-EM map and a map calculated from the fitted model; the red line is the FSC between maps from independent halves of the data. Dotted lines indicate the 0.5 FSC criterion for the map/model comparison and 0.143 for the half datasets. The cryo-EM map was filtered to 3.36 Å, causing the map-to-model correlation to drop to 0 at that resolution. DOI:
http://dx.doi.org/10.7554/eLife.01963.014
Figure 11.. Polypeptide sequence and secondary structure.
α-helices are highlighted in green, β-strands in blue. The second line shows a consensus sequence of the protein families (Mills et al., 2013) with fully conserved amino acids in capitals and partly conserved residues in lower case (h: hydrophobic; s: small [GAS]; l: large [LIFYHW]; a: aromatic [FYWH]; z: T or S; n: negative, D or E; p: positive, R or K). (A) FrhA. In the consensus sequence, the [NiFe] ligands are highlighted in orange and the ligands of the third ion in red. (B) FrhG. Ligands of the proximal, medial, and distal [4Fe4S] cluster are shown in yellow, orange, and red, respectively. The cysteines coordinating a putative zinc ion on the FrhG dimer interface are highlighted in magenta. (C) FrhB. The residues for coordination of the iron–sulphur cluster and FAD are highlighted in green and cyan, respectively, and residues lining the F420 access channel in yellow. DOI:
http://dx.doi.org/10.7554/eLife.01963.015
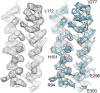
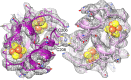
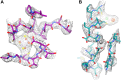
Figure 12.. The ferredoxin domains (residue 206-260) of an FrhG dimer containing the medial and distal FeS clusters.
The two protomers are shown in shades of purple. A high density on the dimer axis between two copies of Cys206 and Cys208 is interpreted as an ion (grey sphere), most likely Zn2+. The ion is ∼9 Å away from the medial FeS cluster (top left and bottom right). DOI:
http://dx.doi.org/10.7554/eLife.01963.017
Figure 13.. FAD cofactor in FrhB with part of its binding pocket.
Conserved residues are labelled in black. Other residues mentioned in the text are grey. (A) The phosphate moiety (orange) sits in a pocket formed by A23–T30 at the C-terminal end of helix 28–39. The adenine moiety (left) is coordinated by the loop A72–N81. (B) In this view, the loop I132–F138 surrounding the isoalloxazine ring can be seen as well as the highly conserved loop G73–T77. DOI:
http://dx.doi.org/10.7554/eLife.01963.018
Figure 14.. Cartoon of the FrhABG heterotrimer (top) and the tetrahedral complex of 12 trimers.
FrhA is green, FrhG magenta, and FrhB blue. The [NiFe] center in FrhA is shown as green and orange spheres, the three [4Fe4S] clusters as orange and yellow spheres, and the FAD in FrhB as a stick model with yellow carbons, the ion in FrhA is orange, and the putative zinc ion on the twofold axis of the FrhG dimer is grey. DOI:
http://dx.doi.org/10.7554/eLife.01963.022
Figure 15.
(A) The proximal FeS cluster of FrhG is coordinated by three cysteine residues and an aspartate (Asp60) with clear density. (B) An ion in FrhA (grey sphere) is coordinated by the C-terminal His386, the main chain oxygen of Ala347 and by Glu44, one of the few carboxylate residues with clear density. DOI:
http://dx.doi.org/10.7554/eLife.01963.023
Similar articles
- De novo modeling of the F(420)-reducing [NiFe]-hydrogenase from a methanogenic archaeon by cryo-electron microscopy.
Mills DJ, Vitt S, Strauss M, Shima S, Vonck J. Mills DJ, et al. Elife. 2013 Mar 5;2:e00218. doi: 10.7554/eLife.00218. Elife. 2013. PMID: 23483797 Free PMC article. - The F₄₂₀-reducing [NiFe]-hydrogenase complex from Methanothermobacter marburgensis, the first X-ray structure of a group 3 family member.
Vitt S, Ma K, Warkentin E, Moll J, Pierik AJ, Shima S, Ermler U. Vitt S, et al. J Mol Biol. 2014 Jul 29;426(15):2813-26. doi: 10.1016/j.jmb.2014.05.024. Epub 2014 Jun 2. J Mol Biol. 2014. PMID: 24887099 - Isolation of an H2-dependent electron-bifurcating CO2-reducing megacomplex with MvhB polyferredoxin from Methanothermobacter marburgensis.
Nomura S, Paczia N, Kahnt J, Shima S. Nomura S, et al. FEBS J. 2024 Jun;291(11):2449-2460. doi: 10.1111/febs.17115. Epub 2024 Mar 12. FEBS J. 2024. PMID: 38468562 - Cryo-EM Structure Determination Using Segmented Helical Image Reconstruction.
Fromm SA, Sachse C. Fromm SA, et al. Methods Enzymol. 2016;579:307-28. doi: 10.1016/bs.mie.2016.05.034. Epub 2016 Jun 28. Methods Enzymol. 2016. PMID: 27572732 Review. - Functional diversity of prokaryotic HdrA(BC) modules: Role in flavin-based electron bifurcation processes and beyond.
Appel L, Willistein M, Dahl C, Ermler U, Boll M. Appel L, et al. Biochim Biophys Acta Bioenerg. 2021 Apr 1;1862(4):148379. doi: 10.1016/j.bbabio.2021.148379. Epub 2021 Jan 16. Biochim Biophys Acta Bioenerg. 2021. PMID: 33460586 Review.
Cited by
- Validation methods for low-resolution fitting of atomic structures to electron microscopy data.
Xu XP, Volkmann N. Xu XP, et al. Arch Biochem Biophys. 2015 Sep 1;581:49-53. doi: 10.1016/j.abb.2015.06.017. Epub 2015 Jun 24. Arch Biochem Biophys. 2015. PMID: 26116787 Free PMC article. - Building Protein Atomic Models from Cryo-EM Density Maps and Residue Co-Evolution.
Bouvier G, Bardiaux B, Pellarin R, Rapisarda C, Nilges M. Bouvier G, et al. Biomolecules. 2022 Sep 13;12(9):1290. doi: 10.3390/biom12091290. Biomolecules. 2022. PMID: 36139128 Free PMC article. - De novo protein structure determination from near-atomic-resolution cryo-EM maps.
Wang RY, Kudryashev M, Li X, Egelman EH, Basler M, Cheng Y, Baker D, DiMaio F. Wang RY, et al. Nat Methods. 2015 Apr;12(4):335-8. doi: 10.1038/nmeth.3287. Epub 2015 Feb 23. Nat Methods. 2015. PMID: 25707029 Free PMC article. - Oxygen-Sensitive Metalloprotein Structure Determination by Cryo-Electron Microscopy.
Cherrier MV, Vernède X, Fenel D, Martin L, Arragain B, Neumann E, Fontecilla-Camps JC, Schoehn G, Nicolet Y. Cherrier MV, et al. Biomolecules. 2022 Mar 12;12(3):441. doi: 10.3390/biom12030441. Biomolecules. 2022. PMID: 35327633 Free PMC article. - CryoEM structures of membrane pore and prepore complex reveal cytolytic mechanism of Pneumolysin.
van Pee K, Neuhaus A, D'Imprima E, Mills DJ, Kühlbrandt W, Yildiz Ö. van Pee K, et al. Elife. 2017 Mar 21;6:e23644. doi: 10.7554/eLife.23644. Elife. 2017. PMID: 28323617 Free PMC article.
References
- Alex LA, Reeve JN, Orme-Johnson WH, Walsh CT. 1990. Cloning, sequence determination, and expression of the genes encoding the subunits of the nickel-containing 8-hydroxy-5-deazaflavin reducing hydrogenase from Methanobacterium thermoautotrophicum deltaH. Biochemistry 29:7237–7244. 10.1021/bi00483a011 - DOI - PubMed
- Armache J-P, Jarasch A, Anger AM, Villa E, Becker T, Bhushan S, Jossinet F, Habeck M, Dindar GL, Franckenberg S, Marquez V, Mielke T, Thomm M, Berninghausen O, Beatrix B, Söding J, Westhof E, Wilson DN, Beckmann R. 2010. Cryo-EM structure and rRNA model of a translating eukaryotic 80S ribosome at 5.5-Å resolution. Proceedings of the National Academy of Sciences of the United States of America 107:19748–19753. 10.1073/pnas.1009999107 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources