The stoichiometry of the nucleoporin 62 subcomplex of the nuclear pore in solution - PubMed (original) (raw)
The stoichiometry of the nucleoporin 62 subcomplex of the nuclear pore in solution
Alexander Ulrich et al. Mol Biol Cell. 2014 May.
Abstract
The nuclear pore complex (NPC) regulates transport between the nucleus and cytoplasm. Soluble cargo-protein complexes navigate through the pore by binding to phenylalanine-glycine (FG)-repeat proteins attached to the channel walls. The Nup62 complex contains the FG-repeat proteins Nup62, Nup54, and Nup58 and is located in the center of the NPC. The three proteins bind each other via conserved coiled-coil segments. To determine the stoichiometry of the Nup62 complex, we undertook an in vitro study using gel filtration and analytical ultracentrifugation. Our results reveal a 1:1:1 stoichiometry of the Nup62 complex, where Nup54 is central with direct binding to Nup62 and Nup58. At high protein concentration, the complex forms larger assemblies while maintaining the Nup62:Nup54:Nup58 ratio. For the homologous Nsp1 complex from Saccharomyces cerevisiae, we determine the same stoichiometry, indicating evolutionary conservation. Furthermore, we observe that eliminating one binding partner can result in the formation of complexes with noncanonical stoichiometry, presumably because unpaired coiled-coil elements tend to find a promiscuous binding partner. We suggest that these noncanonical stoichiometries observed in vitro are unlikely to be physiologically relevant.
Figures
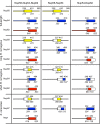
FIGURE 1:
Protein complexes used in this study. Definitions of all Nup58-Nup54-Nup62, Nup58-Nup54, and Nup54-Nup62 complexes used in our study. Narrow tubes represent FG-repeat regions; broad cylinders represent α-helical, predicted coiled-coil domains. Numbers indicate residue positions in the R. norvegicus proteins. Numbers in italics indicate protein residues in the S. cerevisiae homologues. Regions present in the particular complex are colored. The scNup49-Nup57-T4 Lysozyme complex contains, in addition to the residues of the scNup49-Nup57 complex, T4 Lysozyme N-terminally fused to Nup57.
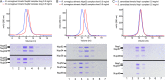
FIGURE 2:
Final purity of protein complexes used in AUC experiments. Size exclusion chromatograms and SDS–PAGE gels of protein complexes used in AUC experiments. Proteins were purified as described in Materials and Methods. Final purity as obtained after gel filtration with S200 10/300 column of (A) Nup58-Nup54-Nup62 complexes, (B) Nup58-Nup54 complexes, and (C) Nup54-Nup62 complexes, shown by size exclusion chromatograms and SDS–PAGE gels. Nup57 autodegradation fragments are indicated with asterisks.
FIGURE 3:
Concentration-dependent oligomerization of Nup58-Nup54-Nup62 complexes. Size exclusion chromatograms and SDS–PAGE gels of a high (5 mg/ml or 3.8 mg/ml) and a low (0.5 mg/ml) concentration of (A) the long rnNup58-Nup54-Nup62 complex, (B) the short rnNup58-Nup54-Nup62 complex, and (C) the scNup49-Nup57-Nsp1 complex. scNup57 autodegradation fragments are indicated by asterisks. Experiments were performed as described in Materials and Methods.
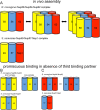
FIGURE 4:
Models of protein subcomplexes of the Nup62/Nsp1 complex. Complex models are based on size exclusion chromatography and AUC data. Cylinders symbolize predicted coiled-coil portions of nucleoporins. Double arrows indicate dynamic exchange between oligomeric states. Cartoon representations of oligomerization behavior and composition of (A) the trimeric complexes, (B) the rnNup54-Nup58/scNup49-Nup57 two-component complexes, and (C) the rnNup54-Nup62/scNup57-Nsp1 two-component complexes. We suggest that complexes depicted in (A) likely represent the complex ensemble in vivo, whereas complexes depicted in (B) and (C) represent assemblies, which, likely artificially, occur in vitro in the absence of the third binding partner.
Similar articles
- Ordered Regions of Channel Nucleoporins Nup62, Nup54, and Nup58 Form Dynamic Complexes in Solution.
Sharma A, Solmaz SR, Blobel G, Melčák I. Sharma A, et al. J Biol Chem. 2015 Jul 24;290(30):18370-8. doi: 10.1074/jbc.M115.663500. Epub 2015 May 29. J Biol Chem. 2015. PMID: 26025361 Free PMC article. - The Nup62 Coiled-Coil Motif Provides Plasticity for Triple-Helix Bundle Formation.
Dewangan PS, Sonawane PJ, Chouksey AR, Chauhan R. Dewangan PS, et al. Biochemistry. 2017 Jun 6;56(22):2803-2811. doi: 10.1021/acs.biochem.6b01050. Epub 2017 Apr 26. Biochemistry. 2017. PMID: 28406021 - Crystal structure of the metazoan Nup62•Nup58•Nup54 nucleoporin complex.
Chug H, Trakhanov S, Hülsmann BB, Pleiner T, Görlich D. Chug H, et al. Science. 2015 Oct 2;350(6256):106-10. doi: 10.1126/science.aac7420. Epub 2015 Aug 20. Science. 2015. PMID: 26292704 - The Quest for the Blueprint of the Nuclear Pore Complex.
Glavy JS. Glavy JS. Protein J. 2019 Aug;38(4):363-376. doi: 10.1007/s10930-019-09858-z. Protein J. 2019. PMID: 31410705 Free PMC article. Review. - Lighting up the nuclear pore complex.
Kahms M, Hüve J, Wesselmann R, Farr JC, Baumgärtel V, Peters R. Kahms M, et al. Eur J Cell Biol. 2011 Sep;90(9):751-8. doi: 10.1016/j.ejcb.2011.04.004. Epub 2011 May 31. Eur J Cell Biol. 2011. PMID: 21632146 Review.
Cited by
- Ordered Regions of Channel Nucleoporins Nup62, Nup54, and Nup58 Form Dynamic Complexes in Solution.
Sharma A, Solmaz SR, Blobel G, Melčák I. Sharma A, et al. J Biol Chem. 2015 Jul 24;290(30):18370-8. doi: 10.1074/jbc.M115.663500. Epub 2015 May 29. J Biol Chem. 2015. PMID: 26025361 Free PMC article. - The Nuclear Pore Complex as a Flexible and Dynamic Gate.
Knockenhauer KE, Schwartz TU. Knockenhauer KE, et al. Cell. 2016 Mar 10;164(6):1162-1171. doi: 10.1016/j.cell.2016.01.034. Cell. 2016. PMID: 26967283 Free PMC article. Review. - Linker Nups connect the nuclear pore complex inner ring with the outer ring and transport channel.
Fischer J, Teimer R, Amlacher S, Kunze R, Hurt E. Fischer J, et al. Nat Struct Mol Biol. 2015 Oct;22(10):774-81. doi: 10.1038/nsmb.3084. Epub 2015 Sep 7. Nat Struct Mol Biol. 2015. PMID: 26344569 - Channel nuclear pore complex subunits are required for transposon silencing in Drosophila.
Munafò M, Lawless VR, Passera A, MacMillan S, Bornelöv S, Haussmann IU, Soller M, Hannon GJ, Czech B. Munafò M, et al. Elife. 2021 Apr 15;10:e66321. doi: 10.7554/eLife.66321. Elife. 2021. PMID: 33856346 Free PMC article. - Analysis of the initiation of nuclear pore assembly by ectopically targeting nucleoporins to chromatin.
Schwartz M, Travesa A, Martell SW, Forbes DJ. Schwartz M, et al. Nucleus. 2015;6(1):40-54. doi: 10.1080/19491034.2015.1004260. Nucleus. 2015. PMID: 25602437 Free PMC article.
References
- Alber F, et al. The molecular architecture of the nuclear pore complex. Nature. 2007;450:695–701. - PubMed
- Amlacher S, Sarges P, Flemming D, van Noort V, Kunze R, Devos DP, Arumugam M, Bork P, Hurt E. Insight into structure and assembly of the nuclear pore complex by utilizing the genome of a eukaryotic thermophile. Cell. 2011;146:277–289. - PubMed
- Beck M, Förster F, Ecke M, Plitzko JM, Melchior F, Gerisch G, Baumeister W, Medalia O. Nuclear pore complex structure and dynamics revealed by cryoelectron tomography. Science. 2004;306:1387–1390. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases