A genotype-first approach to defining the subtypes of a complex disease - PubMed (original) (raw)
A genotype-first approach to defining the subtypes of a complex disease
Holly A Stessman et al. Cell. 2014.
Abstract
Medical genetics typically entails the detailed characterization of a patient's phenotypes followed by genotyping to discover the responsible gene or mutation. Here, we propose that the systematic discovery of genetic variants associated with complex diseases such as autism are progressing to a point where a reverse strategy may be fruitful in assigning the pathogenic effects of many different genes and in determining whether particular genotypes manifest as clinically recognizable phenotypes. This "genotype-first" approach for complex disease necessitates the development of large, highly integrated networks of researchers, clinicians, and patient families, with the promise of improved therapies for subsets of patients.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures
Figure 1. Schematic of Genotype-First Approach for ASD
(A–C) Following complex neurodevelopmental disease diagnosis in the clinic, step one is to apply next-generation sequencing (exome or whole-genome) to identify high-impact rare or de novo variants that exist(s) in an individual. Through screening of many individuals, recurrent mutations in a gene or locus are identified with a general diagnosis of ASD or DD. These candidates are selected for targeted resequencing using high-throughput, cost-effective technologies. Molecular inversion probe (MIP) technology, for example, applied to thousands of individuals with ASD or DD identifies genes with an excess mutational burden in probands when compared to controls. Such genes are most likely to contribute to disease etiology and represent future targets for therapeutic intervention. Families with these gene mutations are recontacted and brought back to the clinic for more comprehensive phenotyping (step two). There will be those genes that have a common, strong, single clinical phenotype (A); however, these will likely be rare. There may be those individual genotypes that all affect the same functional pathway (molecular subtypes) and result in similar or potentially opposing phenotypes (e.g., macrocephaly versus microcephaly) (B). Some mutations even within the same gene, however, may have multiple associated clinical phenotypes (C), suggesting high variability in the type of mutation relative to its gene function and/or incomplete penetrance. The latter especially will require more in-depth study for genetic background effects (step three). This approach will group patients foremost based on genotypes or sets of mutated genes. Larger groups of patients with the same presumptive genetic etiology are re-examined to identify specific clinical phenotypes, with the goal of improving diagnosis, patient care, and management.
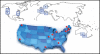
Figure 2. The Autism Spectrum/Intellectual Disability Network
The Autism Spectrum/Intellectual Disability network (ASID), composed of 21 basic research and clinical laboratories from around the world, has assembled >15,000 patients for gene resequencing. The network is broader than the Autism Sequencing Consortium (ASC) in that it considers patients with ASD, ID, epilepsy, or DD due to their comorbidity. It emphasizes collections where parental DNA is available and where patient recontact is possible to accurately resolve phenotype-genotype correlations. The network includes 10 clinical research labs across the world (blue squares) and 11 labs in the USA (red dots) that recruited families as part of the SSC, where patient recontact has now been made possible after extensive IRB review. Subsets of patients with mutations in a common gene are being reassessed to determine whether the mutation defines a clinical subtype.
Figure 3. Potential Genetic Definition of Autism Subtypes
Individuals with autism have been described as having an increased head circumference size (HCZ) distribution (Courchesne et al., 2003). This is observed among ASD probands of the SSC where HCZ is positively skewed (blue arrow) when compared to a normal distribution (left). Subselecting patients (red) with de novo mutations in the β-catenin/Wnt-signaling network (Iossifov et al., 2012; Neale et al., 2012; O’Roak et al., 2012b; Sanders et al., 2012)—defined as those de novo proband events that cluster around the central CTNNB1 node using either STRING or Ingenuity Pathway Analysis enrichment (n = 26 individuals)—further transforms this into a bimodal distribution (right), suggesting reciprocal macrocephaly and microcephaly associated with de novo mutations in this pathway.
Similar articles
- The discovery of integrated gene networks for autism and related disorders.
Hormozdiari F, Penn O, Borenstein E, Eichler EE. Hormozdiari F, et al. Genome Res. 2015 Jan;25(1):142-54. doi: 10.1101/gr.178855.114. Epub 2014 Nov 5. Genome Res. 2015. PMID: 25378250 Free PMC article. - 5-HTTLPR Genotype-Specific Phenotype in Children and Adolescents With Autism.
Brune CW, Kim SJ, Salt J, Leventhal BL, Lord C, Cook EH Jr. Brune CW, et al. Am J Psychiatry. 2006 Dec;163(12):2148-56. doi: 10.1176/ajp.2006.163.12.2148. Am J Psychiatry. 2006. PMID: 17151167 - Heritable genotype contrast mining reveals novel gene associations specific to autism subgroups.
Spencer M, Takahashi N, Chakraborty S, Miles J, Shyu CR. Spencer M, et al. J Biomed Inform. 2018 Jan;77:50-61. doi: 10.1016/j.jbi.2017.11.016. Epub 2017 Nov 29. J Biomed Inform. 2018. PMID: 29197649 Free PMC article. - The genetics of autism.
Muhle R, Trentacoste SV, Rapin I. Muhle R, et al. Pediatrics. 2004 May;113(5):e472-86. doi: 10.1542/peds.113.5.e472. Pediatrics. 2004. PMID: 15121991 Review. - Autism genetics: strategies, challenges, and opportunities.
O'Roak BJ, State MW. O'Roak BJ, et al. Autism Res. 2008 Feb;1(1):4-17. doi: 10.1002/aur.3. Autism Res. 2008. PMID: 19360646 Review.
Cited by
- Decoding mutational hotspots in human disease through the gene modules governing thymic regulatory T cells.
Raposo AASF, Rosmaninho P, Silva SL, Paço S, Brazão ME, Godinho-Santos A, Tokunaga-Mizoro Y, Nunes-Cabaço H, Serra-Caetano A, Almeida ARM, Sousa AE. Raposo AASF, et al. Front Immunol. 2024 Oct 15;15:1458581. doi: 10.3389/fimmu.2024.1458581. eCollection 2024. Front Immunol. 2024. PMID: 39483472 Free PMC article. - Exploration of the intracellular chiral metabolome in pediatric BCP-ALL: a pilot study investigating the metabolic phenotype of IgH locus aberrations.
Collins M, Gorgoglione R, Impedovo V, Pan X, Chakkarai S, Yi SS, Lodi A, Tiziani S. Collins M, et al. Front Oncol. 2024 Aug 5;14:1413264. doi: 10.3389/fonc.2024.1413264. eCollection 2024. Front Oncol. 2024. PMID: 39161381 Free PMC article. - A clustering approach identifies an Autism Spectrum Disorder subtype more responsive to chronic oxytocin treatment.
Zhao W, Le J, Liu Q, Zhu S, Lan C, Zhang Q, Zhang Y, Li Q, Kou J, Yang W, Zhang R, Becker B, Zhang L, Kendrick KM. Zhao W, et al. Transl Psychiatry. 2024 Jul 29;14(1):312. doi: 10.1038/s41398-024-03025-4. Transl Psychiatry. 2024. PMID: 39075076 Free PMC article. Clinical Trial. - The Current Progress of Psychiatric Genomics.
Nishioka M. Nishioka M. Juntendo Iji Zasshi. 2022 Feb 16;68(1):2-11. doi: 10.14789/jmj.JMJ21-0038-R. eCollection 2022. Juntendo Iji Zasshi. 2022. PMID: 38911007 Free PMC article. Review. - Shared and divergent mental health characteristics of ADNP-, CHD8- and DYRK1A-related neurodevelopmental conditions.
Neuhaus E, Rea H, Jones E, Benavidez H, Miles C, Whiting A, Johansson M, Eayrs C, Kurtz-Nelson EC, Earl R, Bernier RA, Eichler EE. Neuhaus E, et al. J Neurodev Disord. 2024 Apr 15;16(1):15. doi: 10.1186/s11689-024-09532-1. J Neurodev Disord. 2024. PMID: 38622540 Free PMC article.
References
- American Psychiatric Association . Diagnostic and Statistical Manual of Mental Disorders. Fifth Edition American Pyschiatric Publishing; Arlington, VA: 2013.
- Bitsika V, Sharpley CF, Orapeleng S. An exploratory analysis of the use of cognitive, adaptive and behavioural indices for cluster analysis of ASD subgroups. J. Intellect. Disabil. Res. 2008;52:973–985. - PubMed
- Charman T, Taylor E, Drew A, Cockerill H, Brown JA, Baird G. Outcome at 7 years of children diagnosed with autism at age 2: predictive validity of assessments conducted at 2 and 3 years of age and pattern of symptom change over time. J. Child Psychol. Psychiatry. 2005;46:500–513. - PubMed
- Consortium, V. Simons Vip Consortium Simons Variation in Individuals Project (Simons VIP): a genetics-first approach to studying autism spectrum and related neurodevelopmental disorders. Neuron. 2012;73:1063–1067. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources