The emergence of the Middle East respiratory syndrome coronavirus - PubMed (original) (raw)
Review
The emergence of the Middle East respiratory syndrome coronavirus
Shauna Milne-Price et al. Pathog Dis. 2014 Jul.
Abstract
On September 20, 2012, a Saudi Arabian physician reported the isolation of a novel coronavirus from a patient with pneumonia on ProMED-mail. Within a few days, the same virus was detected in a Qatari patient receiving intensive care in a London hospital, a situation reminiscent of the role air travel played in the spread of severe acute respiratory syndrome coronavirus (SARS-CoV) in 2002. SARS-CoV originated in China's Guangdong Province and affected more than 8000 patients in 26 countries before it was contained 6 months later. Over a year after the emergence of this novel coronavirus--Middle East respiratory syndrome coronavirus (MERS-CoV)--it has caused 178 laboratory-confirmed cases and 76 deaths. The emergence of a second highly pathogenic coronavirus within a decade highlights the importance of a coordinated global response incorporating reservoir surveillance, high-containment capacity with fundamental and applied research programs, and dependable communication pathways to ensure outbreak containment. Here, we review the current state of knowledge on the epidemiology, ecology, molecular biology, clinical features, and intervention strategies of the novel coronavirus, MERS-CoV.
Keywords: MERS-CoV; coronavirus; epidemiology; intervention strategies; molecular biology.
Published 2014. This article is a US Government work and is in the public domain in the USA.
Figures
Figure 1
False‐color
MERS
‐
C
o
V
particle visualized by electron microscopy. A
MERS
‐
C
o
V
particle (yellow) attached to the surface of a cell (red). The characteristic
MERS
‐
C
o
V
spike glycoproteins are clearly visible on the surface of the
MERS
‐
C
o
V
particle.
Figure 2
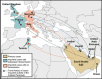
Geographic distribution of the
MERS
‐CoV outbreak. The geographic distribution of
MERS
‐
C
o
V
cases up to February 1, 2014 is shown. Travel history of cases imported outside of the Arabian Peninsula is indicated with dotted arrows. Countries with primary
MERS
‐
C
o
V
cases are shown in brown, countries with imported
MERS
‐
C
o
V
cases and no confirmed human‐to‐human transmission are shown in pink, and countries with imported
MERS
‐
C
o
V
cases and subsequent human‐to‐human transmission are shown in green.
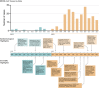
Figure 3
Timeline of the
MERS
‐
C
o
V
outbreak. Temporal distribution of
MERS
‐CoV cases from March 2012 through December 2013. Significant outbreak events and scientific advances during this time period are highlighted below.
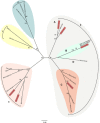
Figure 4
Coronavirus phylogeny. Phylogenetic tree of coronaviruses with representatives of each of the four genera; Alpha (pink), Beta (gray), Delta (blue), and Gammacoronavirus (yellow). Betacoronaviruses are further subdivided into clades A through D, with clade B (green) containing
SARS
‐CoV and clade C (orange) containing
MERS
‐CoV. All known human coronaviruses are represented in red. Maximum‐likelihood trees were generated with the
MEGA
5 software package using a 1231‐nucleotide segment within the RdRp. Trees were visualized using Figtree. Bootstrap values above 75 are shown. CoV isolation origin abbreviations as follows: H: human; Bt: bat; Bt
SL
: bat
SARS
‐like;
BW
: beluga whale;
IBV
: chicken;
FIPV
: feline;
TGEV
: swine; M: mink;
MHV
: murine; Th: thrush; Bu: bulbul; Mun: munia.
Figure 5
Putative
MERS
‐CoV transmission cycle. The putative transmission cycle for
MERS
‐CoV.
MERS
‐CoV likely originated from bats, acting as the natural reservoir. From the natural reservoir,
MERS
‐CoV spilled either directly over to humans (green arrow) or via an intermediate host (dromedary camels, purple arrow). Currently, the exact route of zoonotic transmission of
MERS
‐CoV into the human population remains unknown although the presence of
MERS
‐CoV neutralizing antibodies and the detection of
MERS
‐CoV in dromedary camels suggest that this species is likely to play a major role in the emergence of
MERS
‐CoV. Phylogenetic analysis suggests that multiple introductions of
MERS
‐CoV into the human population have occurred and both zoonotic transmission events and human‐to‐human transmission (blue arrows) drive the current
MERS
‐CoV outbreak.
Figure 6
MERS
‐CoV spike glycoprotein and
DPP
4 receptor interaction. (a) The linear organization of the S1 subunit of
MERS
‐CoV spike glycoprotein with the variable
RBD
located at amino acid residues 367–607, with a
RBM
containing the critical amino acid residues for binding at residues 484–567. (b) The crystal structure of the
MERS
‐CoV
RBD
coupled with the receptor dipeptidyl peptidase
IV
(
DPP
4).
DPP
4 is structurally divided into an alpha‐ and beta‐hydrolase domain, and a beta‐propeller domain. The beta‐propeller domain of
DPP
4 (pink) interacts with the
RBM
region (light blue) of the
MERS
‐CoV spike protein
RBD
. The schematic representation of the
DPP
4 –
MERS
‐CoV spike protein
RBD
structure was generated using chimera and protein accession number 4
KR
0 (Lu et al., 2013).
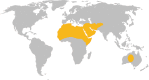
Figure 7
Geographic distribution of dromedary camels. The global distribution of dromedary camels is indicated by yellow shading (Mukasa‐Mugerwa, 1981).
Similar articles
- Middle East Respiratory Syndrome (MERS).
Rasmussen SA, Watson AK, Swerdlow DL. Rasmussen SA, et al. Microbiol Spectr. 2016 Jun;4(3). doi: 10.1128/microbiolspec.EI10-0020-2016. Microbiol Spectr. 2016. PMID: 27337460 Review. - Cross-sectional study of MERS-CoV-specific RNA and antibodies in animals that have had contact with MERS patients in Saudi Arabia.
Kasem S, Qasim I, Al-Hufofi A, Hashim O, Alkarar A, Abu-Obeida A, Gaafer A, Elfadil A, Zaki A, Al-Romaihi A, Babekr N, El-Harby N, Hussien R, Al-Sahaf A, Al-Doweriej A, Bayoumi F, Poon LLM, Chu DKW, Peiris M, Perera RAPM. Kasem S, et al. J Infect Public Health. 2018 May-Jun;11(3):331-338. doi: 10.1016/j.jiph.2017.09.022. Epub 2017 Oct 6. J Infect Public Health. 2018. PMID: 28993171 Free PMC article. - The Middle East Respiratory Syndrome Coronavirus - A Continuing Risk to Global Health Security.
Azhar EI, Lanini S, Ippolito G, Zumla A. Azhar EI, et al. Adv Exp Med Biol. 2017;972:49-60. doi: 10.1007/5584_2016_133. Adv Exp Med Biol. 2017. PMID: 27966107 Free PMC article. - Middle East respiratory syndrome: An emerging coronavirus infection tracked by the crowd.
Mackay IM, Arden KE. Mackay IM, et al. Virus Res. 2015 Apr 16;202:60-88. doi: 10.1016/j.virusres.2015.01.021. Epub 2015 Feb 2. Virus Res. 2015. PMID: 25656066 Free PMC article. Review. - Limited Genetic Diversity Detected in Middle East Respiratory Syndrome-Related Coronavirus Variants Circulating in Dromedary Camels in Jordan.
Seifert SN, Schulz JE, Ricklefs S, Letko M, Yabba E, Hijazeen ZS, Holloway P, Al-Omari B, Talafha HA, Tibbo M, Adney DR, Guitian J, Amarin N, Richt JA, McDowell C, Steel J, Abu-Basha EA, Al-Majali AM, van Doremalen N, Munster VJ. Seifert SN, et al. Viruses. 2021 Mar 31;13(4):592. doi: 10.3390/v13040592. Viruses. 2021. PMID: 33807288 Free PMC article.
Cited by
- Discovery of novel virus sequences in an isolated and threatened bat species, the New Zealand lesser short-tailed bat (Mystacina tuberculata).
Wang J, Moore NE, Murray ZL, McInnes K, White DJ, Tompkins DM, Hall RJ. Wang J, et al. J Gen Virol. 2015 Aug;96(8):2442-2452. doi: 10.1099/vir.0.000158. Epub 2015 Apr 21. J Gen Virol. 2015. PMID: 25900137 Free PMC article. - Peptide-Protein Interaction Studies of Antimicrobial Peptides Targeting Middle East Respiratory Syndrome Coronavirus Spike Protein: An In Silico Approach.
Mustafa S, Balkhy H, Gabere M. Mustafa S, et al. Adv Bioinformatics. 2019 Jul 1;2019:6815105. doi: 10.1155/2019/6815105. eCollection 2019. Adv Bioinformatics. 2019. PMID: 31354813 Free PMC article. - Estimation Of Direct Medical Costs Of Middle East Respiratory Syndrome Coronavirus Infection: A Single-Center Retrospective Chart Review Study.
AlRuthia Y, Somily AM, Alkhamali AS, Bahari OH, AlJuhani RJ, Alsenaidy M, Balkhi B. AlRuthia Y, et al. Infect Drug Resist. 2019 Nov 7;12:3463-3473. doi: 10.2147/IDR.S231087. eCollection 2019. Infect Drug Resist. 2019. PMID: 31819541 Free PMC article. - Middle East respiratory syndrome coronavirus: transmission, virology and therapeutic targeting to aid in outbreak control.
Durai P, Batool M, Shah M, Choi S. Durai P, et al. Exp Mol Med. 2015 Aug 28;47(8):e181. doi: 10.1038/emm.2015.76. Exp Mol Med. 2015. PMID: 26315600 Free PMC article. Review. - An Overview of Antiviral Peptides and Rational Biodesign Considerations.
Lee YJ, Shirkey JD, Park J, Bisht K, Cowan AJ. Lee YJ, et al. Biodes Res. 2022 May 17;2022:9898241. doi: 10.34133/2022/9898241. eCollection 2022. Biodes Res. 2022. PMID: 37850133 Free PMC article. Review.
References
- Albarrak A, Stephens G, Hewson R & Memish Z (2012) Recovery from severe novel coronavirus infection. Saudi Med J 33: 1265–1269. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous