Assessment of four DNA fragments (COI, 16S rDNA, ITS2, 12S rDNA) for species identification of the Ixodida (Acari: Ixodida) - PubMed (original) (raw)
Comparative Study
doi: 10.1186/1756-3305-7-93.
Shaoqiang Wu, Yongning Zhang, Yan Chen, Chunyan Feng, Xiangfen Yuan, Guangle Jia, Junhua Deng, Caixia Wang, Qin Wang, Lin Mei, Xiangmei Lin 1
Affiliations
- PMID: 24589289
- PMCID: PMC3945964
- DOI: 10.1186/1756-3305-7-93
Comparative Study
Assessment of four DNA fragments (COI, 16S rDNA, ITS2, 12S rDNA) for species identification of the Ixodida (Acari: Ixodida)
Jizhou Lv et al. Parasit Vectors. 2014.
Abstract
Background: The 5' region of cytochrome oxidase I (COI) is the standard marker for DNA barcoding. However, COI has proved to be of limited use in identifying some species, and for some taxa, the coding sequence is not efficiently amplified by PCR. These deficiencies lead to uncertainty as to whether COI is the most suitable barcoding fragment for species identification of ticks.
Methods: In this study, we directly compared the relative effectiveness of COI, 16S ribosomal DNA (rDNA), nuclear ribosomal internal transcribed spacer 2 (ITS2) and 12S rDNA for tick species identification. A total of 307 sequences from 84 specimens representing eight tick species were acquired by PCR. Besides the 1,834 published sequences of 189 tick species from GenBank and the Barcode of Life Database, 430 unpublished sequences representing 59 tick species were also successfully screened by Bayesian analyses. Thereafter, the performance of the four DNA markers to identify tick species was evaluated by identification success rates given by these markers using nearest neighbour (NN), BLASTn, liberal tree-based or liberal tree-based (+threshold) methods.
Results: Genetic divergence analyses showed that the intra-specific divergence of each marker was much lower than the inter-specific divergence. Our results indicated that the rates of correct sequence identification for all four markers (COI, 16S rDNA, ITS2, 12S rDNA) were very high (> 96%) when using the NN methodology. We also found that COI was not significantly better than the other markers in terms of its rate of correct sequence identification. Overall, BLASTn and NN methods produced higher rates of correct species identification than that produced by the liberal tree-based methods (+threshold or otherwise).
Conclusions: As the standard DNA barcode, COI should be the first choice for tick species identification, while 16S rDNA, ITS2, and 12S rDNA could be used when COI does not produce reliable results. Besides, NN and BLASTn are efficient methods for species identification of ticks.
Figures
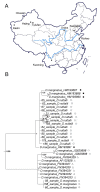
Figure 1
Data collection and phylogenetic screening of sequences. (A) Localities where the 84 tick specimens used in this study were collected. (B) Phylogeny of ticks resulting from Bayesian analysis of COI. Numbers on branches are posterior probabilities. ○, sequence amplified from our specimens; △, previously published sequences; □, unpublished sequences included in data set; ■, misplaced unpublished sequences excluded from data set.
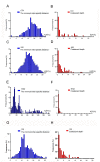
Figure 2
Frequency distributions of K2P distances of COI, 16S rDNA, ITS2 and 12S rDNA within and among tick species. Panels show the distributions of minimum inter-specific K2P distances (A, C, E, G) and of coalescent depths (B, D, F, H) for COI (A, B), 16S rDNA (c, d), ITS2 (E, F) and 12S rDNA (G, H).
Similar articles
- Mitochondrial and nuclear multilocus phylogeny of Rhipicephalus ticks from Kenya.
Kanduma EG, Bishop RP, Githaka NW, Skilton RA, Heyne H, Mwacharo JM. Kanduma EG, et al. Mol Phylogenet Evol. 2019 Nov;140:106579. doi: 10.1016/j.ympev.2019.106579. Epub 2019 Aug 9. Mol Phylogenet Evol. 2019. PMID: 31404610 - Development of a DNA barcoding system for the Ixodida (Acari: Ixodida).
Lv J, Wu S, Zhang Y, Zhang T, Feng C, Jia G, Lin X. Lv J, et al. Mitochondrial DNA. 2014 Apr;25(2):142-9. doi: 10.3109/19401736.2013.792052. Epub 2013 Apr 30. Mitochondrial DNA. 2014. PMID: 23631370 - Prospects of using DNA barcoding for species identification and evaluation of the accuracy of sequence databases for ticks (Acari: Ixodida).
Zhang RL, Zhang B. Zhang RL, et al. Ticks Tick Borne Dis. 2014 Apr;5(3):352-8. doi: 10.1016/j.ttbdis.2014.01.001. Epub 2014 Mar 19. Ticks Tick Borne Dis. 2014. PMID: 24656809 - DNA barcoding mosquitoes: advice for potential prospectors.
Beebe NW. Beebe NW. Parasitology. 2018 Apr;145(5):622-633. doi: 10.1017/S0031182018000343. Epub 2018 Mar 22. Parasitology. 2018. PMID: 29564995 Review. - In support of morphology: Molecular analysis successfully delineates the Afrotropical genus Atylotus (Diptera: Tabanidae) into species.
Williams KA, Snyman LP. Williams KA, et al. Acta Trop. 2023 Jan;237:106725. doi: 10.1016/j.actatropica.2022.106725. Epub 2022 Oct 26. Acta Trop. 2023. PMID: 36309106 Review.
Cited by
- Molecular Analysis of Tick-Borne Bacterial Pathogens from Ticks Infesting Animal Hosts in Kyrgyzstan, 2021.
Kim YJ, Seo JY, Park JS, Kim SY, Aknazarov B, Atabekova N, Lee HI. Kim YJ, et al. Microorganisms. 2024 May 22;12(6):1046. doi: 10.3390/microorganisms12061046. Microorganisms. 2024. PMID: 38930428 Free PMC article. - Patterns of Midichloria infection in avian-borne African ticks and their trans-Saharan migratory hosts.
Di Lecce I, Bazzocchi C, Cecere JG, Epis S, Sassera D, Villani BM, Bazzi G, Negri A, Saino N, Spina F, Bandi C, Rubolini D. Di Lecce I, et al. Parasit Vectors. 2018 Feb 22;11(1):106. doi: 10.1186/s13071-018-2669-z. Parasit Vectors. 2018. PMID: 29471857 Free PMC article. - Does Haemaphysalis bispinosa (Acari: Ixodidae) really occur in China?
Chen Z, Li Y, Ren Q, Liu Z, Luo J, Li K, Guan G, Yang J, Han X, Liu G, Luo J, Yin H. Chen Z, et al. Exp Appl Acarol. 2015 Feb;65(2):249-57. doi: 10.1007/s10493-014-9854-3. Epub 2014 Oct 11. Exp Appl Acarol. 2015. PMID: 25304739 - High degree of mitochondrial gene heterogeneity in the bat tick species Ixodes vespertilionis, I. ariadnae and I. simplex from Eurasia.
Hornok S, Estrada-Peña A, Kontschán J, Plantard O, Kunz B, Mihalca AD, Thabah A, Tomanović S, Burazerović J, Takács N, Görföl T, Estók P, Tu VT, Szőke K, Fernández de Mera IG, de la Fuente J, Takahashi M, Yamauchi T, Takano A. Hornok S, et al. Parasit Vectors. 2015 Sep 17;8:457. doi: 10.1186/s13071-015-1056-2. Parasit Vectors. 2015. PMID: 26382218 Free PMC article. - Francisella-Like Endosymbionts and Rickettsia Species in Local and Imported Hyalomma Ticks.
Azagi T, Klement E, Perlman G, Lustig Y, Mumcuoglu KY, Apanaskevich DA, Gottlieb Y. Azagi T, et al. Appl Environ Microbiol. 2017 Aug 31;83(18):e01302-17. doi: 10.1128/AEM.01302-17. Print 2017 Sep 15. Appl Environ Microbiol. 2017. PMID: 28710265 Free PMC article.
References
- Apanaskevich DA, Horak IG. The genus Hyalomma Koch, 1844: V. Re-evaluation of the taxonomic rank of taxa comprising the H.(Euhyalomma) marginatum Koch complex of species (Acari: Ixodidae) with redescription of all parasitic stages and notes on biology. Int J Acarol. 2008;34(1):13–42. doi: 10.1080/01647950808683704. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials