Community-wide transcriptome of the oral microbiome in subjects with and without periodontitis - PubMed (original) (raw)
Community-wide transcriptome of the oral microbiome in subjects with and without periodontitis
Ana E Duran-Pinedo et al. ISME J. 2014 Aug.
Abstract
Despite increasing knowledge on phylogenetic composition of the human microbiome, our understanding of the in situ activities of the organisms in the community and their interactions with each other and with the environment remains limited. Characterizing gene expression profiles of the human microbiome is essential for linking the role of different members of the bacterial communities in health and disease. The oral microbiome is one of the most complex microbial communities in the human body and under certain circumstances, not completely understood, the healthy microbial community undergoes a transformation toward a pathogenic state that gives rise to periodontitis, a polymicrobial inflammatory disease. We report here the in situ genome-wide transcriptome of the subgingival microbiome in six periodontally healthy individuals and seven individuals with periodontitis. The overall picture of metabolic activities showed that iron acquisition, lipopolysaccharide synthesis and flagellar synthesis were major activities defining disease. Unexpectedly, the vast majority of virulence factors upregulated in subjects with periodontitis came from organisms that are not considered major periodontal pathogens. One of the organisms whose gene expression profile was characterized was the uncultured candidate division TM7, showing an upregulation of putative virulence factors in the diseased community. These data enhance understanding of the core activities that are characteristic of periodontal disease as well as the role that individual organisms in the subgingival community play in periodontitis.
Figures
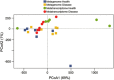
Figure 1
Principal coordinate analysis (PCoA) of phylogenetic profiles from metagenomes and metatranscriptomes. Ordination graph for the first two axes of a PCoA of the phylogenetic profiles obtained from the metagenome and metatranscriptome of the oral microbial communities in healthy sites and sites with severe periodontitis. The % on the axes represents the variance explained by each of the coordinates.
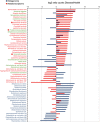
Figure 2
Rank distribution of statistically significant relative increase in number of hits for the metagenome and metatranscriptome results. The ratio of counts in disease vs health was log2 transformed and plotted according to ranks. The statistical significance was calculated using the non-parametric test implemented in the program NOISeq as described in the Materials and methods section. Only species with significant differences, according to NOISeq, either in metagenomic or metatranscriptomic counts, are presented. In green, species with statistical differences in both metagenome and metatranscriptome. In blue, species with statistical differences in metagenomic counts. In red, species with statistical differences in metatranscriptome counts. Red star indicates major periodontal pathogens previously assigned to the ‘red-complex.'
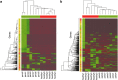
Figure 3
Heat maps of gene expression profiles in the metatranscriptomes. (a) Profiles of the 1212 most highly expressed genes across samples. Genes were first normalized by gene length and _χ_2 transformed before analysis. Most highly expressed genes were selected according to the sum of normalized counts across samples. (b) Profiles of the 1000 most highly differentially expressed genes across samples. Genes were selected from the NOISeq results and counts were _χ_2transformed before analysis. Color bars in red correspond to healthy samples and in green to periodontitis samples.
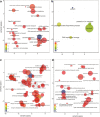
Figure 4
GO enrichment analysis summarized and visualized as a scatter plot using REVIGO. (a) Summarized GO terms related to biological processes in periodontitis. (b) Summarized GO terms related to biological processes in health. (c) Summarized GO terms related to molecular function in periodontitis. (d) Summarized GO terms related to molecular function in health. GO terms are represented by circles and are plotted according to semantic similarities to other GO terms (adjoining circles are most closely related). Circle size is proportional to the frequency of the GO term, whereas color indicates the log10 P value (red higher, blue lower).
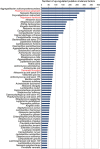
Figure 5
Ranked species by the number of upregulated putative virulence factors in the metatranscriptome. Putative virulence factors were identified by alignment of the protein sequences from the different genomes against the Virulence Factors of Pathogenic Bacteria Database as described in the Materials and methods section. Number in the graph are the absolute number of hits for the different species for the putative virulence factors identified. In red are the members of the ‘red-complex.'
Similar articles
- Small RNA Transcriptome of the Oral Microbiome during Periodontitis Progression.
Duran-Pinedo AE, Yost S, Frias-Lopez J. Duran-Pinedo AE, et al. Appl Environ Microbiol. 2015 Oct;81(19):6688-99. doi: 10.1128/AEM.01782-15. Epub 2015 Jul 17. Appl Environ Microbiol. 2015. PMID: 26187962 Free PMC article. Clinical Trial. - Multi-site analysis of biosynthetic gene clusters from the periodontitis oral microbiome.
Koohi-Moghadam M, Watt RM, Leung WK. Koohi-Moghadam M, et al. J Med Microbiol. 2024 Oct;73(10). doi: 10.1099/jmm.0.001898. J Med Microbiol. 2024. PMID: 39378072 - Deep sequencing of the oral microbiome reveals signatures of periodontal disease.
Liu B, Faller LL, Klitgord N, Mazumdar V, Ghodsi M, Sommer DD, Gibbons TR, Treangen TJ, Chang YC, Li S, Stine OC, Hasturk H, Kasif S, Segrè D, Pop M, Amar S. Liu B, et al. PLoS One. 2012;7(6):e37919. doi: 10.1371/journal.pone.0037919. Epub 2012 Jun 4. PLoS One. 2012. PMID: 22675498 Free PMC article. - Metatranscriptome of the Oral Microbiome in Health and Disease.
Solbiati J, Frias-Lopez J. Solbiati J, et al. J Dent Res. 2018 May;97(5):492-500. doi: 10.1177/0022034518761644. Epub 2018 Mar 8. J Dent Res. 2018. PMID: 29518346 Free PMC article. Review. - The oral microbiome in health and disease.
Wade WG. Wade WG. Pharmacol Res. 2013 Mar;69(1):137-43. doi: 10.1016/j.phrs.2012.11.006. Epub 2012 Nov 28. Pharmacol Res. 2013. PMID: 23201354 Review.
Cited by
- Bacteria associated with periodontal disease are also increased in health.
López-Martínez J, Chueca N, Padial-Molina M, Fernandez-Caballero JA, García F, O'Valle F, Galindo-Moreno P. López-Martínez J, et al. Med Oral Patol Oral Cir Bucal. 2020 Nov 1;25(6):e745-e751. doi: 10.4317/medoral.23766. Med Oral Patol Oral Cir Bucal. 2020. PMID: 32701927 Free PMC article. - A Longitudinal Metagenomic Comparative Analysis of Oral Microbiome Shifts in Patients Receiving Proton Radiation versus Photon Radiation for Head and Neck Cancer.
Meiller TF, Fraser CM, Grant-Beurmann S, Humphrys M, Tallon L, Sadzewicz LD, Jabra-Rizk MA, Alfaifi A, Kensara A, Molitoris JK, Witek M, Mendes WS, Regine WF, Tran PT, Miller RC, Sultan AS. Meiller TF, et al. J Cancer Allied Spec. 2024 Jan 22;10(1):579. doi: 10.37029/jcas.v10i1.579. eCollection 2024. J Cancer Allied Spec. 2024. PMID: 38259673 Free PMC article. - A microbial perspective of human developmental biology.
Charbonneau MR, Blanton LV, DiGiulio DB, Relman DA, Lebrilla CB, Mills DA, Gordon JI. Charbonneau MR, et al. Nature. 2016 Jul 7;535(7610):48-55. doi: 10.1038/nature18845. Nature. 2016. PMID: 27383979 Free PMC article. - A pilot study examining periodontally healthy middle-aged humans and monkeys display different levels of alveolar bone resorption, gingival inflammatory infiltrate, and salivary microbiota profile.
Lin B, Pathak JL, Gao H, Zhou Z, Ser HL, Wu L, Lee LH, Wang L, Chen J, Zhong M. Lin B, et al. PLoS One. 2024 Oct 16;19(10):e0311282. doi: 10.1371/journal.pone.0311282. eCollection 2024. PLoS One. 2024. PMID: 39413077 Free PMC article. - Metagenomics, Metatranscriptomics, and Metabolomics Approaches for Microbiome Analysis.
Aguiar-Pulido V, Huang W, Suarez-Ulloa V, Cickovski T, Mathee K, Narasimhan G. Aguiar-Pulido V, et al. Evol Bioinform Online. 2016 May 12;12(Suppl 1):5-16. doi: 10.4137/EBO.S36436. eCollection 2016. Evol Bioinform Online. 2016. PMID: 27199545 Free PMC article. Review.
References
- Bender MH, Weiser JN. (2006). The atypical amino-terminal LPNTG-containing domain of the pneumococcal human IgA1-specific protease is required for proper enzyme localization and function. Mol Microbiol 61: 526–543. - PubMed
- Booijink CCGM, Boekhorst J, Zoetendal EG, Smidt H, Kleerebezem M, de Vos WM. (2010). Metatranscriptome analysis of the human fecal microbiota reveals subject-specific expression profiles, with genes encoding proteins involved in carbohydrate metabolism being dominantly expressed. Appl Environ Microbiol 76: 5533–5540. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources