DAWN: a framework to identify autism genes and subnetworks using gene expression and genetics - PubMed (original) (raw)
doi: 10.1186/2040-2392-5-22.
Jing Lei, Stephan J Sanders, Arthur Jeremy Willsey, Yan Kou, Abdullah Ercument Cicek, Lambertus Klei, Cong Lu, Xin He, Mingfeng Li, Rebecca A Muhle, Avi Ma'ayan, James P Noonan, Nenad Sestan, Kathryn A McFadden, Matthew W State, Joseph D Buxbaum, Bernie Devlin, Kathryn Roeder 1
Affiliations
- PMID: 24602502
- PMCID: PMC4016412
- DOI: 10.1186/2040-2392-5-22
DAWN: a framework to identify autism genes and subnetworks using gene expression and genetics
Li Liu et al. Mol Autism. 2014.
Abstract
Background: De novo loss-of-function (dnLoF) mutations are found twofold more often in autism spectrum disorder (ASD) probands than their unaffected siblings. Multiple independent dnLoF mutations in the same gene implicate the gene in risk and hence provide a systematic, albeit arduous, path forward for ASD genetics. It is likely that using additional non-genetic data will enhance the ability to identify ASD genes.
Methods: To accelerate the search for ASD genes, we developed a novel algorithm, DAWN, to model two kinds of data: rare variations from exome sequencing and gene co-expression in the mid-fetal prefrontal and motor-somatosensory neocortex, a critical nexus for risk. The algorithm casts the ensemble data as a hidden Markov random field in which the graph structure is determined by gene co-expression and it combines these interrelationships with node-specific observations, namely gene identity, expression, genetic data and the estimated effect on risk.
Results: Using currently available genetic data and a specific developmental time period for gene co-expression, DAWN identified 127 genes that plausibly affect risk, and a set of likely ASD subnetworks. Validation experiments making use of published targeted resequencing results demonstrate its efficacy in reliably predicting ASD genes. DAWN also successfully predicts known ASD genes, not included in the genetic data used to create the model.
Conclusions: Validation studies demonstrate that DAWN is effective in predicting ASD genes and subnetworks by leveraging genetic and gene expression data. The findings reported here implicate neurite extension and neuronal arborization as risks for ASD. Using DAWN on emerging ASD sequence data and gene expression data from other brain regions and tissues would likely identify novel ASD genes. DAWN can also be used for other complex disorders to identify genes and subnetworks in those disorders.
Figures
Figure 1
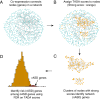
The DAWN algorithm. (A) Each node in the network represents a gene and each edge represents pairs of genes with strong co-expression (absolute correlation _r_>0.7). (B) Orange nodes indicate genes with strong genetic scores from the TADA test. (C) Hot spots (i.e., clusters of strong scores) are classified as nASD genes in the screening stage of the algorithm; cool spots (i.e. strong scores in isolation) are not.(D) In the final cleaning step, the nASD list is further refined to reveal the rASD gene list. This step uses the TADA scores and features of the network to compute the false discovery rate of each gene. FDR, false discovery rate; nASD, network autism spectrum disorder; rASD, risk autism spectrum disorder; TADA, transmission and de novo association.
Figure 2
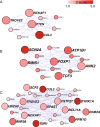
Network of risk ASD (rASD) genes. These genes met the false discovery rate threshold of.05. The intensity of the red reflects the magnitude of the netscore based on TADA statistics from neighboring genes. Large nodes depict genes that have at least one dnLoF mutation. CUL3, DYRK1A, GRIN2B, POGZ, SCN2A and TBR1 are genes with multiple dnLoF mutations. Edges connect genes with absolute correlation of.7 or greater based on periods 3–5 or 4–6. dnLoF, de novo loss of function; rASD, risk autism spectrum disorder; TADA, transmission and de novo association.
Figure 3
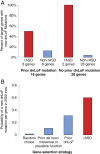
Analysis of MIPS validation experiment. (A) First 44 genes with prior de novo mutations were sequenced for 2,448 additional trios. These genes were cross-classified by whether or not they had a prior dnLoF mutation, and whether or not they were on the DAWN rASD list (yes: red, no: blue). For each category, the percentage of genes that had a dnLoF mutation in the new trios is depicted.(B) For a given gene, the probability of observing a dnLoF mutation in 2,500 probands varies. This probability is compared for four types of genes: a randomly chosen gene and three classifications of the genes from the MIPS experiment including: (i) all 44 genes, (ii) those 16 genes with a prior dnLoF mutation and (iii) those 10 genes on the rASD list. dnLoF, de novo loss of function; rASD, risk autism spectrum disorder.
Figure 4
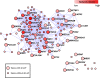
Clustering by enrichment and protein-protein interaction (PPI). The rASD genes are seeded into the PPI network presented in [6], represented by red nodes, with size proportional to the number of connections. The blue nodes are immediate intermediate proteins [36]. The network was clustered using organic clustering methods implemented in yEd [44] rASD, risk autism spectrum disorder.
Figure 5
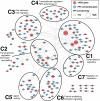
Gene subnetworks for the PTEN , FOXP1 and SPAST genes. Figure shows all rASD genes with absolute correlation of.7 or better with (A)PTEN, (B)FOXP1 and (C)SPAST. Intensity of red color reflects the magnitude of the _Z_-score from the TADA statistic. Large nodes with labels depict genes that have at least one dnLoF mutation recorded in the current literature (except PTEN). ANK2, CUL3, DYRK1A, SCN2A and TBR1 are genes with multiple dnLoF mutations. ANK2 is included along with rASD genes because it has two dnLoF mutations.
Figure 6
Predicted risk genes functionally related to neuron outgrowth. (A) Ten rASD genes are GO-identified for neuron outgrowth and 16 additional genes rASD genes are directly connected via the PPI network. (B) Note that 68 rASD genes are either GO-identified for neuron outgrowth or separated in the PPI network from genes annotated by neurite outgrowth by at most one step. PPI, protein-protein interaction; rASD, risk autism spectrum disorder.
Similar articles
- Mutations and Modeling of the Chromatin Remodeler CHD8 Define an Emerging Autism Etiology.
Barnard RA, Pomaville MB, O'Roak BJ. Barnard RA, et al. Front Neurosci. 2015 Dec 17;9:477. doi: 10.3389/fnins.2015.00477. eCollection 2015. Front Neurosci. 2015. PMID: 26733790 Free PMC article. Review. - Leveraging blood serotonin as an endophenotype to identify de novo and rare variants involved in autism.
Chen R, Davis LK, Guter S, Wei Q, Jacob S, Potter MH, Cox NJ, Cook EH, Sutcliffe JS, Li B. Chen R, et al. Mol Autism. 2017 Mar 21;8:14. doi: 10.1186/s13229-017-0130-3. eCollection 2017. Mol Autism. 2017. PMID: 28344757 Free PMC article. - Targeted sequencing and functional analysis reveal brain-size-related genes and their networks in autism spectrum disorders.
Li J, Wang L, Guo H, Shi L, Zhang K, Tang M, Hu S, Dong S, Liu Y, Wang T, Yu P, He X, Hu Z, Zhao J, Liu C, Sun ZS, Xia K. Li J, et al. Mol Psychiatry. 2017 Sep;22(9):1282-1290. doi: 10.1038/mp.2017.140. Epub 2017 Jul 25. Mol Psychiatry. 2017. PMID: 28831199 - Functional relationships between recessive inherited genes and genes with de novo variants in autism spectrum disorder.
Wang L, Zhang Y, Li K, Wang Z, Wang X, Li B, Zhao G, Fang Z, Ling Z, Luo T, Xia L, Li Y, Guo H, Hu Z, Li J, Sun Z, Xia K. Wang L, et al. Mol Autism. 2020 Oct 6;11(1):75. doi: 10.1186/s13229-020-00382-x. Mol Autism. 2020. PMID: 33023636 Free PMC article. - The genetics of autism.
Muhle R, Trentacoste SV, Rapin I. Muhle R, et al. Pediatrics. 2004 May;113(5):e472-86. doi: 10.1542/peds.113.5.e472. Pediatrics. 2004. PMID: 15121991 Review.
Cited by
- De novo Mutations (DNMs) in Autism Spectrum Disorder (ASD): Pathway and Network Analysis.
Alonso-Gonzalez A, Rodriguez-Fontenla C, Carracedo A. Alonso-Gonzalez A, et al. Front Genet. 2018 Sep 21;9:406. doi: 10.3389/fgene.2018.00406. eCollection 2018. Front Genet. 2018. PMID: 30298087 Free PMC article. Review. - Forecasting risk gene discovery in autism with machine learning and genome-scale data.
Brueggeman L, Koomar T, Michaelson JJ. Brueggeman L, et al. Sci Rep. 2020 Mar 12;10(1):4569. doi: 10.1038/s41598-020-61288-5. Sci Rep. 2020. PMID: 32165711 Free PMC article. - Association of CDH11 with Autism Spectrum Disorder Revealed by Matched-gene Co-expression Analysis and Mouse Behavioral Studies.
Wu N, Wang Y, Jia JY, Pan YH, Yuan XB. Wu N, et al. Neurosci Bull. 2022 Jan;38(1):29-46. doi: 10.1007/s12264-021-00770-0. Epub 2021 Sep 14. Neurosci Bull. 2022. PMID: 34523068 Free PMC article. - Mutations and Modeling of the Chromatin Remodeler CHD8 Define an Emerging Autism Etiology.
Barnard RA, Pomaville MB, O'Roak BJ. Barnard RA, et al. Front Neurosci. 2015 Dec 17;9:477. doi: 10.3389/fnins.2015.00477. eCollection 2015. Front Neurosci. 2015. PMID: 26733790 Free PMC article. Review. - Integrative analysis of transcriptome dynamics during human craniofacial development identifies candidate disease genes.
Yankee TN, Oh S, Winchester EW, Wilderman A, Robinson K, Gordon T, Rosenfeld JA, VanOudenhove J, Scott DA, Leslie EJ, Cotney J. Yankee TN, et al. Nat Commun. 2023 Aug 2;14(1):4623. doi: 10.1038/s41467-023-40363-1. Nat Commun. 2023. PMID: 37532691 Free PMC article.
References
- Buxbaum JD, Daly MJ, Devlin B, Lehner T, Roeder K, State MW, Barrett J, Bilder D, Boerwinkle E, Brudno M, Burbach P, Buxbaum JD, Camp N, Chahrour M, Cook EH, Coon H, Coppola G, Coulter M, Cutler D, Daly MJ, dePristo M, Devlin B, Eichler EE, Fromer M, Geschwind DH, Gibbs RA, Gill M, Goldberg AP, Haines JL. et al.The autism sequencing consortium: large-scale, high-throughput sequencing in autism spectrum disorders. Neuron. 2012;76(6):1052–1056. doi: 10.1016/j.neuron.2012.12.008. - DOI - PMC - PubMed
- Sanders SJ, Ercan-Sencicek AG, Hus V, Luo R, Murtha MT, Moreno-De-Luca D, Chu SH, Moreau MP, Gupta AR, Thomson SA, Mason CE, Bilguvar K, Celestino-Soper PB, Choi M, Crawford EL, Davis L, Wright NR, Dhodapkar RM, DiCola M, DiLullo NM, Fernandez TV, Fielding-Singh V, Fishman DO, Frahm S, Garagaloyan R, Goh GS, Kammela S, Klei L, Lowe JK, Lund SC. et al.Multiple recurrent de novo , CNVs, including duplications of the 7q11.23 Williams syndrome region, are strongly associated with autism. Neuron. 2011;70(5):863–885. doi: 10.1016/j.neuron.2011.05.002. - DOI - PMC - PubMed
- Pinto D, Pagnamenta AT, Klei L, Anney R, Merico D, Regan R, Conroy J, Magalhaes TR, Correia C, Abrahams BS, Almeida J, Bacchelli E, Bader GD, Bailey AJ, Baird G, Battaglia A, Berney T, Bolshakova N, Bolte S, Bolton PF, Bourgeron T, Brennan S, Brian J, Bryson SE, Carson AR, Casallo G, Casey J, Chung BH, Cochrane L. et al.Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2010;466(7304):368–372. doi: 10.1038/nature09146. - DOI - PMC - PubMed
- Iossifov I, Ronemus M, Levy D, Wang Z, Hakker I, Rosenbaum J, Yamrom B, Lee YH, Narzisi G, Leotta A, Kendall J, Grabowska E, Ma B, Marks S, Rodgers L, Stepansky A, Troge J, Andrews P, Bekritsky M, Pradhan K, Ghiban E, Kramer M, Parla J, Demeter R, Fulton LL, Fulton RS, Magrini VJ, Ye K, Darnell JC, Darnell RB. et al. De novo gene disruptions in children on the autistic spectrum. Neuron. 2012;74(2):285–299. doi: 10.1016/j.neuron.2012.04.009. - DOI - PMC - PubMed
- Kong A, Frigge ML, Masson G, Besenbacher S, Sulem P, Magnusson G, Gudjonsson SA, Sigurdsson A, Jonasdottir A, Jonasdottir A, Wong WS, Sigurdsson G, Walters GB, Steinberg S, Helgason H, Thorleifsson G, Gudbjartsson DF, Helgason A, Magnusson OT, Thorsteinsdottir U, Stefansson K. Rate of de novo mutations and the importance of father’s age to disease risk. Nature. 2012;488(7412):471–475. doi: 10.1038/nature11396. - DOI - PMC - PubMed
Grants and funding
- R01 GM094780/GM/NIGMS NIH HHS/United States
- R25 MH077823/MH/NIMH NIH HHS/United States
- U01 MH100239/MH/NIMH NIH HHS/United States
- T32 MH018268/MH/NIMH NIH HHS/United States
- U01 MH100233/MH/NIMH NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources