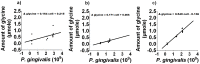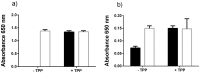Porphyromonas gingivalis and Treponema denticola exhibit metabolic symbioses - PubMed (original) (raw)
Porphyromonas gingivalis and Treponema denticola exhibit metabolic symbioses
Kheng H Tan et al. PLoS Pathog. 2014.
Abstract
Porphyromonas gingivalis and Treponema denticola are strongly associated with chronic periodontitis. These bacteria have been co-localized in subgingival plaque and demonstrated to exhibit symbiosis in growth in vitro and synergistic virulence upon co-infection in animal models of disease. Here we show that during continuous co-culture a P. gingivalis:T. denticola cell ratio of 6∶1 was maintained with a respective increase of 54% and 30% in cell numbers when compared with mono-culture. Co-culture caused significant changes in global gene expression in both species with altered expression of 184 T. denticola and 134 P. gingivalis genes. P. gingivalis genes encoding a predicted thiamine biosynthesis pathway were up-regulated whilst genes involved in fatty acid biosynthesis were down-regulated. T. denticola genes encoding virulence factors including dentilisin and glycine catabolic pathways were significantly up-regulated during co-culture. Metabolic labeling using 13C-glycine showed that T. denticola rapidly metabolized this amino acid resulting in the production of acetate and lactate. P. gingivalis may be an important source of free glycine for T. denticola as mono-cultures of P. gingivalis and T. denticola were found to produce and consume free glycine, respectively; free glycine production by P. gingivalis was stimulated by T. denticola conditioned medium and glycine supplementation of T. denticola medium increased final cell density 1.7-fold. Collectively these data show P. gingivalis and T. denticola respond metabolically to the presence of each other with T. denticola displaying responses that help explain enhanced virulence of co-infections.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
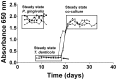
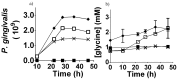
Figure 1. Continuous culture of P. gingivalis and T. denticola mono- and co-cultures.
Cell density of P. gingivalis and T. denticola mono- and co-cultures from three independent continuous cultures in OBGM with the dilution rate of 0.044 h−1 and mean generation time of 15.8 h as determined by measuring A650 nm. The arrow shows the addition of P. gingivalis to a steady state T. denticola culture.
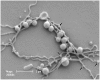
Figure 2. Scanning electron micrograph of P. gingivalis and T. denticola grown in continuous culture.
Co-culture was collected, fixed on a coverslip, dehydrated, covered with colloidal silver, gold-coated and imaged using a Philips XL30 field-emission scanning electron microscope. Electron micrographs showed that P. gingivalis and T. denticola coaggregated. T. denticola is a long helical shaped spirochete with an average length of 5 to 20 µm. P. gingivalis is a coccobacillus with an average diameter of 1 µm. Putative T. denticola outer sheath vesicles and/or P. gingivalis outer membrane vesicles are indicated by arrows along the length of T. denticola.
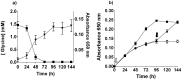
Figure 3. T. denticola growth and glycine.
(a) The concentration of free glycine in T. denticola culture (black square; left axis). T. denticola growth curve in the same medium (black inverted triangle; right axis). Data points are the mean and standard deviation of three biological replicates. (b) Glycine (10 mM) was added to OBGM either before inoculation with T. denticola (black square, open arrow) or at 96 h after inoculation (black triangle, filled arrow) and bacterial growth was determined by A650 nm measurement. T. denticola culture with no added glycine (white circle). Results are expressed as mean ± standard deviation obtained from eight replicates.
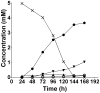
Figure 4. The extracellular products of T. denticola [U-13C]glycine fermentation.
[U-13C]glycine (5.00 mM) was added to a 24 h T. denticola culture and aliquots were collected every 24 h, filtered and the identity and the quantity of the 13C-labeled compounds was determined using NMR spectroscopy. black cross, [U-13C]glycine; black circle, [U-13C]acetate; black inverted triangle, dual or uniformly-labeled lactate; black triangle, [U-13C]bicarbonate; white circle, dual or uniformly-labeled alanine.
Figure 5. P. gingivalis cell numbers and free glycine content in different cultures.
a) The cell numbers of P. gingivalis in different media as determined by absorbance at 650 nm. b) The concentration of free glycine in different P. gingivalis cultures over time, as determined by GC-MS. Data shown are the average of three biological replicates. P. gingivalis grown in:- OBGM – black diamond; OBGM/PBS – black cross; OBGM/T. denticola conditioned medium – white square. Uninoculated OBGM/T. denticola conditioned medium – black square.
Figure 6. Free glycine production during P. gingivalis growth.
The difference in the amount of free glycine relative to that at t = 0 h as a function of P. gingivalis cell numbers in a) OBGM/PBS, b) OBGM and c) OBGM/T. denticola conditioned medium. A regression line was fitted using a linear mixed modelling approach. The slope represents the amount of glycine produced/109 P. gingivalis cells.
Figure 7. Excess thiamine pyrophosphate production by P. gingivalis.
The E. coli TPP auxotrophic strain JW3957-1 (black shading) and the parent strain JRG902 (white shading) were cultured in M9 growth medium (that lacks thiamine) supplemented with either (a) uninoculated P. gingivalis medium (that lacks thiamine) or (b) cell-free P. gingivalis spent medium. The bacterium was cultured with or without TPP addition (5.88 nM).
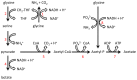
Figure 8. Proposed T. denticola glycine catabolic pathways.
Glycine can be oxidized by the glycine cleavage system (1), producing NH3, CO2 and CH2-THF. Glycine and CH2-THF can be condensed to form serine by serine hydroxymethyltransferase (2). Serine is deaminated to produce pyruvate by serine dehydratase (3). Lactate dehydrogenase (4) catalyzes the interconversion of pyruvate and lactate with concomitant interconversion of NADH and NAD+. Pyruvate can also be metabolized to acetate by pyruvate-ferredoxin oxidoreductase (5), phosphate acetyltransferase (6) and acetate kinase (7). Glycine can also be reduced to acetyl-P by the glycine reductase system (8).
Similar articles
- Metabolic cooperativity between Porphyromonas gingivalis and Treponema denticola.
Kin LX, Butler CA, Slakeski N, Hoffmann B, Dashper SG, Reynolds EC. Kin LX, et al. J Oral Microbiol. 2020 Aug 24;12(1):1808750. doi: 10.1080/20002297.2020.1808750. J Oral Microbiol. 2020. PMID: 32944158 Free PMC article. - The Role of Treponema denticola Motility in Synergistic Biofilm Formation With Porphyromonas gingivalis.
Ng HM, Slakeski N, Butler CA, Veith PD, Chen YY, Liu SW, Hoffmann B, Dashper SG, Reynolds EC. Ng HM, et al. Front Cell Infect Microbiol. 2019 Dec 18;9:432. doi: 10.3389/fcimb.2019.00432. eCollection 2019. Front Cell Infect Microbiol. 2019. PMID: 31921707 Free PMC article. - [Pathogenic potential of Porphyromonas gingivalis, Treponema denticola and Tannerella forsythia, the red bacterial complex associated with periodontitis].
Bodet C, Chandad F, Grenier D. Bodet C, et al. Pathol Biol (Paris). 2007 Apr-May;55(3-4):154-62. doi: 10.1016/j.patbio.2006.07.045. Epub 2006 Oct 17. Pathol Biol (Paris). 2007. PMID: 17049750 Review. French. - [Research progress in the interaction between Treponema denticola and microorganisms of subgingival plaque].
Huang PE, Shen DN, Wu YF. Huang PE, et al. Zhonghua Kou Qiang Yi Xue Za Zhi. 2021 Apr 9;56(4):380-384. doi: 10.3760/cma.j.cn112144-20200528-00305. Zhonghua Kou Qiang Yi Xue Za Zhi. 2021. PMID: 33832042 Review. Chinese.
Cited by
- Porphyromonas gingivalis interaction with Candida albicans allows for aerobic escape, virulence and adherence.
de Jongh CA, Bikker FJ, de Vries TJ, Werner A, Gibbs S, Krom BP. de Jongh CA, et al. Biofilm. 2023 Dec 17;7:100172. doi: 10.1016/j.bioflm.2023.100172. eCollection 2024 Jun. Biofilm. 2023. PMID: 38226024 Free PMC article. - Metabolic Signaling and Spatial Interactions in the Oral Polymicrobial Community.
Miller DP, Fitzsimonds ZR, Lamont RJ. Miller DP, et al. J Dent Res. 2019 Nov;98(12):1308-1314. doi: 10.1177/0022034519866440. Epub 2019 Jul 29. J Dent Res. 2019. PMID: 31356756 Free PMC article. Review. - Periodontal Disease: The Good, The Bad, and The Unknown.
Sedghi LM, Bacino M, Kapila YL. Sedghi LM, et al. Front Cell Infect Microbiol. 2021 Dec 7;11:766944. doi: 10.3389/fcimb.2021.766944. eCollection 2021. Front Cell Infect Microbiol. 2021. PMID: 34950607 Free PMC article. Review. - Periopathogenic bacteria in dental plaque of Congolese patients with periodontitis: A pilot study.
Kalala-Kazadi E, Sekele-Issouradi JP, Bolenge-Ileboso J, Lasserre JF, Mantshumba-Milolo A, Ntumba-Mulumba H, Brecx MC. Kalala-Kazadi E, et al. J Clin Exp Dent. 2018 Mar 1;10(3):e232-e236. doi: 10.4317/jced.54613. eCollection 2018 Mar. J Clin Exp Dent. 2018. PMID: 29721223 Free PMC article. - Chronic periodontitis and community-acquired pneumonia: a population-based cohort study.
Kim SJ, Kim K, Choi S, Chang J, Kim SM, Park SM, Cho HJ. Kim SJ, et al. BMC Pulm Med. 2019 Dec 30;19(1):268. doi: 10.1186/s12890-019-1017-1. BMC Pulm Med. 2019. PMID: 31888597 Free PMC article.
References
- Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL (1998) Microbial complexes in subgingival plaque. J Clin Periodontol 25: 134–144. - PubMed
- Darveau RP (2010) Periodontitis: a polymicrobial disruption of host homeostasis. Nat Rev Microbiol 8: 481–490. - PubMed
- Marsh PD (2003) Are dental diseases examples of ecological catastrophes? Microbiology 149: 279–294. - PubMed
Publication types
MeSH terms
Grants and funding
This work was supported by the Australian National Health and Medical Research Council Project Grant Number 1008061. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
LinkOut - more resources
Full Text Sources
Other Literature Sources