A human short open reading frame (sORF)-encoded polypeptide that stimulates DNA end joining - PubMed (original) (raw)
A human short open reading frame (sORF)-encoded polypeptide that stimulates DNA end joining
Sarah A Slavoff et al. J Biol Chem. 2014.
Abstract
The recent discovery of numerous human short open reading frame (sORF)-encoded polypeptides (SEPs) has raised important questions about the functional roles of these molecules in cells. Here, we show that a 69-amino acid SEP, MRI-2, physically interacts with the Ku heterodimer to stimulate DNA double-strand break ligation via nonhomologous end joining. The characterization of MRI-2 suggests that this SEP may participate in DNA repair and underscores the potential of SEPs to serve important biological functions in mammalian cells.
Keywords: DNA Repair; Peptide Interactions; Peptides; Protein-Protein Interactions; Proteomics; Short ORF.
Figures
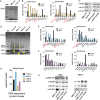
FIGURE 1.
Detection of C7ORF49 isoform 2 (MRI-2) in K562 cells. A, peptidomics workflow to identify nonannotated short ORFs. The K562 cellular peptidome was isolated, fractionated, and subjected to liquid chromatography-mass spectrometry. Nonannotated peptides were identified by SEQUEST search against a K562 RNA deep sequencing library and subsequent removal of annotated sequences. B, MS/MS spectrum of the unique C7ORF49/MRI-2 C-terminal tryptic peptide, with detected fragment ions marked in blue (b ions) and red (y ions). C, multiple sequence alignment (ClustalW2) of the three MRI isoforms reveals that isoforms 1 and 2 have identical N-terminal sequences of 46 amino acids (red) before a frameshift generates a unique C-terminal sequence for isoform 2 (blue for isoform 2, green for isoform 1). Isoform 3 is identical to isoform 1, but lacks the N-terminal 46 amino acids.
FIGURE 2.
MRI-2 interacts with Ku. A, MRI-FLAG was co-immunoprecipitated (IP) from HEK 293T cells and analyzed by SDS-PAGE followed by Coomassie Brilliant Blue staining. The co-IP of MRI-2 (†) enriched two bands (*), which were identified as Ku70 (XRCC6) and Ku80 (XRCC5) by mass spectrometry (data not shown). MW, molecular weight markers. B, semiquantitative proteomics by spectral counting was used to confirm MRI-2 interaction partners. Bands of interest were excised from experimental and control samples by cutting straight across the gel and subjected to in-gel digest and proteomics. The top 10 most abundant proteins in each experimental sample were plotted in terms of spectral counts and compared with the negative control. Ku70 and Ku80 were enriched 3–10-fold. C, confirmation of MRI-2 interaction with Ku70 and Ku80 by Western blotting. FLAG-tagged MRI-2 or empty vector was immunoprecipitated from HEK 293T cells and then analyzed by Western blotting with antibodies against Ku70 and Ku80. Lysates were simultaneously probed to confirm equal loading. Enrichment of Ku70 and Ku80 by the MRI-2 co-IP as compared with vector background was observed. D, co-immunoprecipitation of MRI-1, MRI-2, and MRI-3 (indicated by arrowhead on gel) from HEK293T, with SDS-PAGE analysis and Coomassie Brilliant Blue staining. The four molecular mass regions analyzed were 70, 80, and 450 kDa (bands of interest indicated by *). E and F, semiquantitative proteomics by spectral counting demonstrates enrichment of Ku70 and Ku80 by MRI-1 (E) but not MRI-3 (F). G, spectral counting for the 450-kDa band for all three proteins shows enrichment of DNA-PK by MRI-1. H, co-IP and Western blotting confirm interactions of MRI-1 with Ku and DNA-PK and the absence of interactions with MRI-3. Lysates were analyzed to demonstrate equal loading.
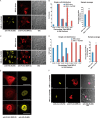
FIGURE 3.
MRI-1–3 nuclear localization requires Ku. A, the subcellular localization of MRI-2 was assessed by transfection into HeLa cells and detection with anti-FLAG immunofluorescence (red) and confocal microscopy. DIC images were acquired to analyze cell morphology. When transfected alone (top), MRI-2 partitions between the nucleus and cytoplasm; when co-transfected with Ku70 and Ku80 (bottom, Ku80-HA, yellow), the nuclear fraction of MRI-2 increases. B, single cell plots and population averages of the percentage of total MRI-2 immunofluorescence in the nucleus reveal a 38% increase in the presence of Ku (*, p < 0.05). Each data set represents >21 cells from four fields of view. An identical analysis in HEK293T cells (C and D) reveals a stronger enhancement of MRI-2 nuclear localization of 82% in the presence of Ku in this cell line (**, p < 0.005). Each data set represents >24 cells from two fields of view. E, the subnuclear distribution of MRI-2 (anti-FLAG, red) is identical to that of co-transfected Ku80 (anti-HA, yellow). F, top, MRI-1 (anti-FLAG, red) is also recruited to the nucleus by Ku co-expression (Ku70-Myc, yellow, and Ku80-HA, pink), but MRI-3 is not (bottom). Therefore, nuclear localization of MRI-1 and -2 may be mediated by their interaction with Ku. All scale bars, 20 μm. Statistical analysis was by two-tailed Student's t test, and error bars represent S.E.
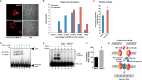
FIGURE 4.
MRI-2 nuclear localization during NHEJ, binding to Ku-DNA complexes, and stimulation of NHEJ in vitro support its functional role in DSB repair. A, representative images of HeLa cells expressing MRI-2 treated either with vehicle (top) or with 50 μ
m
etoposide (bottom) for 4 h. This drug treatment elicits DSBs and activates NHEJ. B and C, single cell plots (B) and population averages (C) of the percentage of total MRI-2 in the nucleus demonstrate an etoposide-mediated increase of 31% (*, p < 0.05). Statistical analysis was by two-tailed Student's t test, and error bars represent S.E. Each data set represents 21 cells from five fields of view. D, MRI-2 interaction with a Ku-DNA complex was assayed by electrophoretic mobility shift. Purified Ku70 and Ku80 were preincubated with a radiolabeled DNA duplex, and increasing amounts of purified MRI-2 were added. Complexes of one and two Ku heterodimers bound to DNA are resolved by gel electrophoresis, and both exhibit progressive, dose-dependent supershift due to binding of multiple copies of MRI-2. E, NHEJ activity was assayed by adding radiolabeled double-stranded DNA probe to NHEJ-competent cell lysates and assaying formation of ligation products (dimers and trimers, bracket) by separation on a gel (top) in the absence or presence of 5 μ
m
purified MRI-2. Wort, Wortmannin. F, quantitation of end-joining activity in four replicate experiments shows an increase in NHEJ efficiency (percentage of DNA ends joined) of 68% as compared with the basal reaction (p = 0.02, Student's t test). G, model of the potential interaction(s) between MRI-2 and the NHEJ machinery. The Ku heterodimer binds to DSBs and recruits other NHEJ factors. MRI-2 binds to Ku-DNA to stimulate this process.
Similar articles
- Detection and repair of ionizing radiation-induced DNA double strand breaks: new developments in nonhomologous end joining.
Wang C, Lees-Miller SP. Wang C, et al. Int J Radiat Oncol Biol Phys. 2013 Jul 1;86(3):440-9. doi: 10.1016/j.ijrobp.2013.01.011. Epub 2013 Feb 20. Int J Radiat Oncol Biol Phys. 2013. PMID: 23433795 Free PMC article. Review. - Synthesis of Biotinylated Inositol Hexakisphosphate To Study DNA Double-Strand Break Repair and Affinity Capture of IP6-Binding Proteins.
Jiao C, Summerlin M, Bruzik KS, Hanakahi L. Jiao C, et al. Biochemistry. 2015 Oct 20;54(41):6312-22. doi: 10.1021/acs.biochem.5b00642. Epub 2015 Oct 7. Biochemistry. 2015. PMID: 26397942 - APLF (C2orf13) facilitates nonhomologous end-joining and undergoes ATM-dependent hyperphosphorylation following ionizing radiation.
Macrae CJ, McCulloch RD, Ylanko J, Durocher D, Koch CA. Macrae CJ, et al. DNA Repair (Amst). 2008 Feb 1;7(2):292-302. doi: 10.1016/j.dnarep.2007.10.008. DNA Repair (Amst). 2008. PMID: 18077224 - Specificity of the dRP/AP lyase of Ku promotes nonhomologous end joining (NHEJ) fidelity at damaged ends.
Strande N, Roberts SA, Oh S, Hendrickson EA, Ramsden DA. Strande N, et al. J Biol Chem. 2012 Apr 20;287(17):13686-93. doi: 10.1074/jbc.M111.329730. Epub 2012 Feb 23. J Biol Chem. 2012. PMID: 22362780 Free PMC article. - Identification and characterization of sORF-encoded polypeptides.
Chu Q, Ma J, Saghatelian A. Chu Q, et al. Crit Rev Biochem Mol Biol. 2015 Mar-Apr;50(2):134-41. doi: 10.3109/10409238.2015.1016215. Epub 2015 Apr 10. Crit Rev Biochem Mol Biol. 2015. PMID: 25857697 Free PMC article. Review.
Cited by
- Microprotein-encoding RNA regulation in cells treated with pro-inflammatory and pro-fibrotic stimuli.
Pai VJ, Lau CJ, Garcia-Ruiz A, Donaldson C, Vaughan JM, Miller B, De Souza EV, Pinto AM, Diedrich J, Gavva NR, Yu S, DeBoever C, Horman SR, Saghatelian A. Pai VJ, et al. BMC Genomics. 2024 Nov 5;25(1):1034. doi: 10.1186/s12864-024-10948-1. BMC Genomics. 2024. PMID: 39497054 Free PMC article. - Evaluation of Eukaryotic mRNA Coding Potential.
Kochetov AV. Kochetov AV. Methods Mol Biol. 2025;2859:319-331. doi: 10.1007/978-1-0716-4152-1_18. Methods Mol Biol. 2025. PMID: 39436610 - Detection of host cell microprotein impurities in antibody drug products.
Tzani I, Castro-Rivadeneyra M, Kelly P, Strasser L, Zhang L, Clynes M, Karger BL, Barron N, Bones J, Clarke C. Tzani I, et al. Nat Commun. 2024 Oct 4;15(1):8605. doi: 10.1038/s41467-024-51870-0. Nat Commun. 2024. PMID: 39366928 Free PMC article. - Deciphering the ghost proteome in ovarian cancer cells by deep proteogenomic characterization.
Garcia-Del Rio DF, Derhourhi M, Bonnefond A, Leblanc S, Guilloy N, Roucou X, Eyckerman S, Gevaert K, Salzet M, Cardon T. Garcia-Del Rio DF, et al. Cell Death Dis. 2024 Sep 30;15(9):712. doi: 10.1038/s41419-024-07046-1. Cell Death Dis. 2024. PMID: 39349928 Free PMC article. - Distinct functions of PAXX and MRI during chromosomal end joining.
Cisneros-Aguirre M, Lopezcolorado FW, Ping X, Chen R, Stark JM. Cisneros-Aguirre M, et al. bioRxiv [Preprint]. 2024 Aug 22:2024.08.21.607864. doi: 10.1101/2024.08.21.607864. bioRxiv. 2024. PMID: 39229097 Free PMC article. Preprint.
References
- Andrews S. J., Rothnagel J. A. (2014) Emerging evidence for functional peptides encoded by short open reading frames. Nat. Rev. Genet. 15, 193–204 - PubMed
- Menschaert G., Van Criekinge W., Notelaers T., Koch A., Crappé J., Gevaert K., Van Damme P. (2013) Deep proteome coverage based on ribosome profiling aids mass spectrometry-based protein and peptide discovery and provides evidence of alternative translation products and near-cognate translation initiation events. Mol. Cell. Proteomics 12, 1780–1790 - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- F32 GM099408/GM/NIGMS NIH HHS/United States
- R01 GM102491/GM/NIGMS NIH HHS/United States
- 1F32GM099408/GM/NIGMS NIH HHS/United States
- R01GM102491/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous