Every amino acid matters: essential contributions of histone variants to mammalian development and disease - PubMed (original) (raw)
Review
Every amino acid matters: essential contributions of histone variants to mammalian development and disease
Ian Maze et al. Nat Rev Genet. 2014 Apr.
Abstract
Despite a conserved role for histones as general DNA packaging agents, it is now clear that another key function of these proteins is to confer variations in chromatin structure to ensure dynamic patterns of transcriptional regulation in eukaryotes. The incorporation of histone variants is particularly important to this process. Recent knockdown and knockout studies in various cellular systems, as well as direct mutational evidence from human cancers, now suggest a crucial role for histone variant regulation in processes as diverse as differentiation and proliferation, meiosis and nuclear reprogramming. In this Review, we provide an overview of histone variants in the context of their unique functions during mammalian germ cell and embryonic development, and examine the consequences of aberrant histone variant regulation in human disease.
Conflict of interest statement
Competing interests statement
The authors declare no competing interests.
Figures
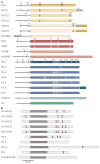
Figure 1. Human core and linker histone variants
a | Variants of the core histones H2A (yellow), H2B (red), H3 (blue) and H4 (green) are shown. Unstructured amino- terminal tails are shown as black lines. Specific amino acid residues are depicted at key differences among variants of a common histone protein family (for example, H2A.X, H2BE and H3.3). Different shades of colour are used to indicate protein sequences that are highly divergent between canonical histones (which are listed first) and their variants (for example, H2A.Bbd, histone H2B type W-T (H2BFWT) and histone H3-like centromeric protein A (CENP-A)). b | Human linker histone variants are shown. Owing to high sequence divergence between variants, specific amino acid differences are not shown. Unstructured amino- and carboxy-terminal domains are shown in light grey. Globular domains are shown in brown. Canonical serine/threonine PXK phosphorylation sites that are targeted by cyclin-dependent kinases are indicated in magenta. Alternative names of variants are given in parentheses. aa, amino acid; mH2A1, macroH2A1. Part a is adapted, with permission, from REF. © (2006) Canadian Science Publishing and has been updated to reflect current data on histone variant structures.
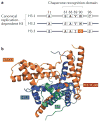
Figure 2. Structural characterization of H3.3 in complex with DAXX: implications for chaperone- specific histone variant deposition
a | Amino acid sequence substitutions of the histone variant H3.3 relative to the canonical replication-dependent histones H3.1 and H3.2 are shown. H3.3 differs from H3.1 and H3.2 at five and four amino acid positions, respectively. b | The crystal structures of the histone-binding domain of death domain-associated protein 6 (DAXX) in complex with a histone H3.3–H4 dimer have recently been solved, which elucidates the principles that underlie the specificity of DAXX-mediated H3.3 recognition. Further functional studies indicated a crucial role for G90 in H3.3 (which forms part of the substituted H3.3 core AAIG motif (FIG. 1)) in DAXX-mediated H3.3 recognition. Data from REF. .
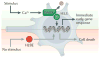
Figure 3. Histone variant exchange in post-replicative cells
Recent findings have suggested distinct mechanisms of histone variant regulation in postmitotic neurons during periods of heightened or reduced cellular activity. In the presence of a stimulus, calcium-dependent dephosphorylation of death domain-associated protein 6 (DAXX) in embryonic cortical neurons leads to an increase in H3.3 deposition at regulatory elements that are associated with activity-dependent immediate early gene responses. Conversely, during periods of reduced sensory experience, expression of H2BE (which is enriched exclusively in olfactory chemosensory neurons) is enhanced, which results in cell death.
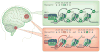
Figure 4. H3.3 variant mutations in brain gliomas: direct evidence that ‘every amino acid matters’
The wild-type histone H3 recruits Polycomb repressive complex 2 (PRC2) and stimulates methyltransferase activity of its catalytic subunit EZH2 (green box),which trimethylates histone H3 at lysine 27 (H3K27me3). The replication-independent histone variant H3.3 mutant that contains the K27M substitution (red box) was recently identified in many diffuse intrinsic pontine gliomas (red circle, bottom left) and supratentorial glioblastomas (red circle, top left). This mutation leads to dominant inhibition of EZH2 in both cis and trans and to concomitant global loss of H3K27me3. These data provide the first direct evidence that mutations in histone variants themselves contribute to human disease.
Similar articles
- New functions for an old variant: no substitute for histone H3.3.
Elsaesser SJ, Goldberg AD, Allis CD. Elsaesser SJ, et al. Curr Opin Genet Dev. 2010 Apr;20(2):110-7. doi: 10.1016/j.gde.2010.01.003. Epub 2010 Feb 12. Curr Opin Genet Dev. 2010. PMID: 20153629 Free PMC article. Review. - Genomic insights into chromatin reprogramming to totipotency in embryos.
Ladstätter S, Tachibana K. Ladstätter S, et al. J Cell Biol. 2019 Jan 7;218(1):70-82. doi: 10.1083/jcb.201807044. Epub 2018 Sep 26. J Cell Biol. 2019. PMID: 30257850 Free PMC article. Review. - Role of H1 linker histones in mammalian development and stem cell differentiation.
Pan C, Fan Y. Pan C, et al. Biochim Biophys Acta. 2016 Mar;1859(3):496-509. doi: 10.1016/j.bbagrm.2015.12.002. Epub 2015 Dec 13. Biochim Biophys Acta. 2016. PMID: 26689747 Free PMC article. Review. - Histone Variant H3.3: A versatile H3 variant in health and in disease.
Xiong C, Wen Z, Li G. Xiong C, et al. Sci China Life Sci. 2016 Mar;59(3):245-56. doi: 10.1007/s11427-016-5006-9. Epub 2016 Jan 29. Sci China Life Sci. 2016. PMID: 26825948 Review. - DNA methylation reprogramming and DNA repair in the mouse zygote.
Lepikhov K, Wossidlo M, Arand J, Walter J. Lepikhov K, et al. Int J Dev Biol. 2010;54(11-12):1565-74. doi: 10.1387/ijdb.103206kl. Int J Dev Biol. 2010. PMID: 21404179
Cited by
- Epigenetic regulation in the inner ear and its potential roles in development, protection, and regeneration.
Layman WS, Zuo J. Layman WS, et al. Front Cell Neurosci. 2015 Jan 7;8:446. doi: 10.3389/fncel.2014.00446. eCollection 2014. Front Cell Neurosci. 2015. PMID: 25750614 Free PMC article. Review. - A histone H3K9M mutation traps histone methyltransferase Clr4 to prevent heterochromatin spreading.
Shan CM, Wang J, Xu K, Chen H, Yue JX, Andrews S, Moresco JJ, Yates JR, Nagy PL, Tong L, Jia S. Shan CM, et al. Elife. 2016 Sep 20;5:e17903. doi: 10.7554/eLife.17903. Elife. 2016. PMID: 27648579 Free PMC article. - Epigenome Maintenance in Response to DNA Damage.
Dabin J, Fortuny A, Polo SE. Dabin J, et al. Mol Cell. 2016 Jun 2;62(5):712-27. doi: 10.1016/j.molcel.2016.04.006. Mol Cell. 2016. PMID: 27259203 Free PMC article. Review. - Quantitative Mass Spectrometry Reveals Changes in Histone H2B Variants as Cells Undergo Inorganic Arsenic-Mediated Cellular Transformation.
Rea M, Jiang T, Eleazer R, Eckstein M, Marshall AG, Fondufe-Mittendorf YN. Rea M, et al. Mol Cell Proteomics. 2016 Jul;15(7):2411-22. doi: 10.1074/mcp.M116.058412. Epub 2016 May 11. Mol Cell Proteomics. 2016. PMID: 27169413 Free PMC article. - MeCP2 and the enigmatic organization of brain chromatin. Implications for depression and cocaine addiction.
Ausió J. Ausió J. Clin Epigenetics. 2016 May 21;8:58. doi: 10.1186/s13148-016-0214-5. eCollection 2016. Clin Epigenetics. 2016. PMID: 27213019 Free PMC article. Review.
References
- Skene PJ, Henikoff S. Histone variants in pluripotency and disease. Development. 2013;140:2513–2524. - PubMed
- Chen P, Zhao J, Li G. Histone variants in development and diseases. J Genet Genom. 2013;40:355–365. - PubMed
- Filipescu D, Szenker E, Almouzni G. Developmental roles of histone H3 variants and their chaperones. Trends Genet. 2013;29:630–640. - PubMed
- Talbert PB, Henikoff S. Histone variants — ancient wrap artists of the epigenome. Nature Rev Mol Cell Biol. 2010;11:264–275. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P50 MH096890/MH/NIMH NIH HHS/United States
- R01 MH094698/MH/NIMH NIH HHS/United States
- P50 MH096890‑01/MH/NIMH NIH HHS/United States
- R01 MH094698‑01/MH/NIMH NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources