The i-AAA protease YME1L and OMA1 cleave OPA1 to balance mitochondrial fusion and fission - PubMed (original) (raw)
The i-AAA protease YME1L and OMA1 cleave OPA1 to balance mitochondrial fusion and fission
Ruchika Anand et al. J Cell Biol. 2014.
Abstract
Mitochondrial fusion and structure depend on the dynamin-like GTPase OPA1, whose activity is regulated by proteolytic processing. Constitutive OPA1 cleavage by YME1L and OMA1 at two distinct sites leads to the accumulation of both long and short forms of OPA1 and maintains mitochondrial fusion. Stress-induced OPA1 processing by OMA1 converts OPA1 completely into short isoforms, inhibits fusion, and triggers mitochondrial fragmentation. Here, we have analyzed the function of different OPA1 forms in cells lacking YME1L, OMA1, or both. Unexpectedly, deletion of Oma1 restored mitochondrial tubulation, cristae morphogenesis, and apoptotic resistance in cells lacking YME1L. Long OPA1 forms were sufficient to mediate mitochondrial fusion in these cells. Expression of short OPA1 forms promoted mitochondrial fragmentation, which indicates that they are associated with fission. Consistently, GTPase-inactive, short OPA1 forms partially colocalize with ER-mitochondria contact sites and the mitochondrial fission machinery. Thus, OPA1 processing is dispensable for fusion but coordinates the dynamic behavior of mitochondria and is crucial for mitochondrial integrity and quality control.
Figures
Figure 1.
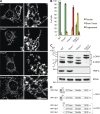
Loss of OMA1 restores tubular mitochondria in _Yme1l_−/− cells. (A) Representative images of mitochondrial morphology in MEFs. The asterisks denote the area of magnification enlarged on the right. Bars: (left) 15 µm; (right) 5 µm. (B) Quantification of three independent experiments (error bars indicate mean ± SD), n ≥ 100. (C) Accumulation of OPA1 forms in MEFs lacking YME1L, OMA1, or both. Cells lysates were analyzed by SDS-PAGE analysis and immunoblotting using the indicated antibodies. a–e, OPA1 forms. (D) Schematic representation of mature L-OPA1 forms derived from splice variants 1 and 7 and S-OPA1 forms produced by cleavage at proteolytic sites S1 or S2 by OMA1 or YME1L, respectively.
Figure 2.
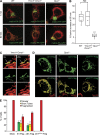
OPA1 processing is dispensable for mitochondrial fusion. (A and B) Photoactivatable GFP (matrix-PA-GFP) and mito-mCherry both targeted to the mitochondrial matrix were expressed in WT, _Yme1l_−/−_Oma1_−/−, and _Opa1_−/− MEFs (A). Fusion was monitored by the time-dependent dilution and redistribution of PA-GFP fluorescence (top, 1 min; bottom, 60 min). Bars, 15 µm. (B) Quantification of mitochondrial fusion. Results are represented in the form of a box plot: boxes represent data between the 25th and 75th percentiles, lines extend between the 10th and 90th percentiles, the horizontal line indicates the median, and the circle indicates the mean value (n ≥ 14; _Yme1l_−/−_Oma1_−/− vs. WT, P = 0.8; _Yme1l_−/−_Oma1_−/− vs. _Opa1_−/−, P = 1.6 × 10−12; WT vs. _Opa1_−/−, P = 4.6 × 10−8; ***, P ≤ 0.001). (C) Visualization of individual fusion events in _Yme1l_−/−_Oma1_−/− MEFs using matrix-PA-GFP and mito-mCherry. The arrowheads indicate mitochondria that have acquired photoactivated GFP due to fusion with photoactivated mitochondria. Bar, 5 µm. (D) Mitochondrial morphology in _Opa1_−/− MEFs expressing cleavable or noncleavable OPA1 forms. Flag-tagged variants of human OPA1 isoform 1 (S1-Flag), isoform 1 lacking the S1 site (ΔS1-Flag), or ΔS1-FlagK301A were transiently expressed in _Opa1_−/− cells. The mitochondrial network was visualized using a TOM20-specific antibody. Bar, 15 µm. (E) Quantification of three independent experiments (error bars indicate mean ± SD), n ≥ 100.
Figure 3.
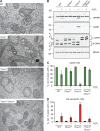
Loss of OMA1 restores cristae morphology and apoptotic resistance of _Yme1l_−/− cells. (A) WT, _Oma1_−/−, _Yme1l_−/−, and _Yme1l_−/−_Oma1_−/− MEFs were analyzed by transmission electron microscopy. Bar, 1 µm. (B–D) WT and protease-deficient MEFs were exposed to H2O2 to induce apoptosis. (B) Immunoblot analysis of apoptotic marker proteins (cleaved PARP, cPARP; caspase 3, CASP). (C and D) Flow cytometry analysis of viable cells (Q3: 7-AAD−;APC−) and late apoptotic cells (Q2: 7-AAD+; APC+). *, P ≤ 0.05; ***, P ≤ 0.001. Error bars indicate mean ± SD.
Figure 4.
The function of short OPA1 forms is linked to mitochondrial fission. (A) Mitochondria fuse in _Yme1_−/− cells. Mitochondrial fusion was analyzed as in Fig. 2 using matrix-PA-GFP. (B) Quantification of mitochondrial fusion in _Yme1l_−/− cells after 60 min. Results are represented in the form of a box plot (see Fig. 2). WT versus _Yme1l_−/− cells, P = 0.14. (C) Expression of S-OPA1 forms in _Yme1l_−/−_Oma1_−/− cells induces mitochondrial fragmentation. Mitochondrial morphology was assessed in _Yme1l_−/−_Oma1_−/− MEFs expressing Flag-tagged variants of rat S-OPA1-Flag or S-OPA1K301A-Flag (bottom) harboring a mutation in the GTPase domain of OPA1. (D) Quantification of three independent experiments (error bars indicate mean ± SD). n ≥ 100; **, P ≥ 0.01. (E) Mitochondrial fusion occurs in fragmented mitochondria of WT and _Yme1l_−/−_Oma1_−/− cells expressing S-OPA1-Flag. (F) Quantification of mitochondrial fusion after 60 min. Results are shown in the form of a box plot (see Fig. 2). Shown are WT/_Yme1l_−/−_Oma1_−/− cells versus WT/_Yme1l_−/−_Oma1_−/− cells expressing rat S-OPA1. Bars, 15 µm.
Figure 5.
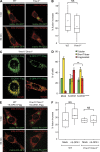
GTPase-inactive short OPA1 forms colocalize with sites of mitochondrial division. (A) S-OPA1K301A-Flag expressed in _Yme1l_−/−_Oma1_−/− cells assembles into punctae partially colocalizing with ER–mitochondria contact sites (i; a total of 90 spots in five different cells), endogenous DRP1 (ii; a total of 230 spots in five different cells), and endogenous MID49 (iii; a total of 186 spots in six different cells). Bars, 2 µm. (B) Line scan of ii and iii along the lines indicated in A. (C) S-OPA1K301A-Flag punctae coalesce at sites of mitochondria constriction. Mitochondria are visualized using mito-mCherry, the ER using GRP78-specific antibodies. Bar, 2 µm.
Similar articles
- Regulation of OPA1 processing and mitochondrial fusion by m-AAA protease isoenzymes and OMA1.
Ehses S, Raschke I, Mancuso G, Bernacchia A, Geimer S, Tondera D, Martinou JC, Westermann B, Rugarli EI, Langer T. Ehses S, et al. J Cell Biol. 2009 Dec 28;187(7):1023-36. doi: 10.1083/jcb.200906084. J Cell Biol. 2009. PMID: 20038678 Free PMC article. - Identification of new OPA1 cleavage site reveals that short isoforms regulate mitochondrial fusion.
Wang R, Mishra P, Garbis SD, Moradian A, Sweredoski MJ, Chan DC. Wang R, et al. Mol Biol Cell. 2021 Jan 15;32(2):157-168. doi: 10.1091/mbc.E20-09-0605. Epub 2020 Nov 25. Mol Biol Cell. 2021. PMID: 33237841 Free PMC article. - Mitochondrial membrane potential and oxidative stress interact to regulate Oma1-dependent processing of Opa1 and mitochondrial dynamics.
Fogo GM, Raghunayakula S, Emaus KJ, Torres Torres FJ, Wider JM, Sanderson TH. Fogo GM, et al. FASEB J. 2024 Sep 30;38(18):e70066. doi: 10.1096/fj.202400313R. FASEB J. 2024. PMID: 39312414 - OPA1 processing in cell death and disease - the long and short of it.
MacVicar T, Langer T. MacVicar T, et al. J Cell Sci. 2016 Jun 15;129(12):2297-306. doi: 10.1242/jcs.159186. Epub 2016 May 17. J Cell Sci. 2016. PMID: 27189080 Review. - OMA1-Mediated Mitochondrial Dynamics Balance Organellar Homeostasis Upstream of Cellular Stress Responses.
Gilkerson R, Kaur H, Carrillo O, Ramos I. Gilkerson R, et al. Int J Mol Sci. 2024 Apr 22;25(8):4566. doi: 10.3390/ijms25084566. Int J Mol Sci. 2024. PMID: 38674151 Free PMC article. Review.
Cited by
- Ischemic Preconditioning and Postconditioning Protect the Heart by Preserving the Mitochondrial Network.
Ismail NI, Michel NA, Katwadi K, Lim MM, Chan TK, Rahman A, Xu D, Ong SG, Hausenloy DJ, Ong SB. Ismail NI, et al. Biomed Res Int. 2022 Sep 27;2022:6889278. doi: 10.1155/2022/6889278. eCollection 2022. Biomed Res Int. 2022. PMID: 36203484 Free PMC article. - Differentiation activates mitochondrial OPA1 processing in myoblast cell lines.
Kaur H, Carrillo O, Garcia I, Ramos I, St Vallier S, De La Torre P, Lopez A, Keniry M, Bazan D, Elizondo J, Grishma KC, Ann MacMillan-Crow L, Gilkerson R. Kaur H, et al. Mitochondrion. 2024 Sep;78:101933. doi: 10.1016/j.mito.2024.101933. Epub 2024 Jul 8. Mitochondrion. 2024. PMID: 38986925 Free PMC article. - Mitochondrial Diseases Part II: Mouse models of OXPHOS deficiencies caused by defects in regulatory factors and other components required for mitochondrial function.
Iommarini L, Peralta S, Torraco A, Diaz F. Iommarini L, et al. Mitochondrion. 2015 May;22:96-118. doi: 10.1016/j.mito.2015.01.008. Epub 2015 Jan 29. Mitochondrion. 2015. PMID: 25640959 Free PMC article. Review. - Mitochondria from the Outside in: The Relationship Between Inter-Organelle Crosstalk and Mitochondrial Internal Organization.
Friedman JR. Friedman JR. Contact (Thousand Oaks). 2022 Jan-Dec;5:25152564221133267. doi: 10.1177/25152564221133267. Contact (Thousand Oaks). 2022. PMID: 36329759 Free PMC article. - A novel MTORC2-AKT-ROS axis triggers mitofission and mitophagy-associated execution of colorectal cancer cells upon drug-induced activation of mutant KRAS.
Iskandar K, Foo J, Liew AQX, Zhu H, Raman D, Hirpara JL, Leong YY, Babak MV, Kirsanova AA, Armand AS, Oury F, Bellot G, Pervaiz S. Iskandar K, et al. Autophagy. 2024 Jun;20(6):1418-1441. doi: 10.1080/15548627.2024.2307224. Epub 2024 Feb 25. Autophagy. 2024. PMID: 38261660 Free PMC article.
References
- Alexander C., Votruba M., Pesch U.E., Thiselton D.L., Mayer S., Moore A., Rodriguez M., Kellner U., Leo-Kottler B., Auburger G., et al. 2000. OPA1, encoding a dynamin-related GTPase, is mutated in autosomal dominant optic atrophy linked to chromosome 3q28. Nat. Genet. 26:211–215 10.1038/79944 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases