Structural polymorphism in the N-terminal oligomerization domain of NPM1 - PubMed (original) (raw)
Structural polymorphism in the N-terminal oligomerization domain of NPM1
Diana M Mitrea et al. Proc Natl Acad Sci U S A. 2014.
Abstract
Nucleophosmin (NPM1) is a multifunctional phospho-protein with critical roles in ribosome biogenesis, tumor suppression, and nucleolar stress response. Here we show that the N-terminal oligomerization domain of NPM1 (Npm-N) exhibits structural polymorphism by populating conformational states ranging from a highly ordered, folded pentamer to a highly disordered monomer. The monomer-pentamer equilibrium is modulated by posttranslational modification and protein binding. Phosphorylation drives the equilibrium in favor of monomeric forms, and this effect can be reversed by Npm-N binding to its interaction partners. We have identified a short, arginine-rich linear motif in NPM1 binding partners that mediates Npm-N oligomerization. We propose that the diverse functional repertoire associated with NPM1 is controlled through a regulated unfolding mechanism signaled through posttranslational modifications and intermolecular interactions.
Keywords: NMR; X-ray crystallography.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
Domain organization of NPM1. NPM1 is composed of an N-terminal oligomerization domain (blue), containing two nuclear export signals (NES), a central disordered region (gray) that incorporates the bipartite nuclear localization signal (NLS), and a C-terminal DNA/RNA binding domain (magenta) harboring the nucleolar localization signal (NoLS). The three evolutionarily conserved acidic tracts, A1, A2, and A3, are represented in red.
Fig. 2.
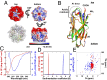
Npm-N converts from a folded pentamer to a disordered monomer as a function of Na+ concentration. (A) Crystal structure of NPM1 N-terminal domain in pentameric form, solved at 1.8 Å. The electrostatic potential map of Npm-N calculated in PyMOL Molecular Graphics System with Adaptive Poisson-Bolzmann Solver (APBS) shows that the N- and C-termini face (Upper) of the pentamer has a large negative net charge, whereas the opposite face (Lower) is charge-neutral. (B) Superimposition of isolated subunits of the pentamer; these contain nine β-strands connected by flexible linkers (residues 13–120). The N- and C-termini are highly flexible and are not observed in the structure. The acidic loop connecting β2 and β3 (A1 tract) exhibits a high degree of flexibility between the otherwise overlapping structures of the five subunits of the pentamer. (C) CD wavelength scans of disordered Npm-N in the absence of NaCl (red) and folded Npm-N in the presence of 0.2 M NaCl (blue). (D) Velocity sedimentation AUC analysis shows the folded state to be pentameric and the disordered state to be monomeric. (E) Two-dimensional transverse relaxation-optimized spectroscopy (TROSY) spectral overlay of the monomeric (red) and pentameric (blue) folds of 2H/15N-labeled Npm-N.
Fig. 3.
Phosphorylation modulates Npm-N oligomerization and folding states. [1H,15N] TROSY spectra of 15N-labeled S48E (A), S88E (B), and T95D (C) highlight the disorder–order landscape of Npm-N generated through phosphomimetic mutations. (D) Location of the phosphorylation sites used in this study to introduce phosphomimetic mutations are represented on two adjacent subunits of Npm-N pentamer. Ser48 (black) and Ser88 (green) are buried at the hydrophobic interface between two protomers, Thr95 (purple) is solvent-exposed, at the tip of the tight loop connecting β6–β6′, and Ser125 (orange), not observed in the crystal structure, is part of acidic tract A2, in the C-terminal flexible tail. (E) The PKA phosphorylation sites are differentially protected from modification, as evidenced by radiolabeled ATP incorporation assays after 30 min incubation at 37 °C. The maximum number of modifications achieved after overnight incubation with PKA for each mutant, as determined by MS, is indicated above the bars (n.d., not detected). The intensity of 32P-radiolabeled gel bands was normalized relative to a histone H1 control, as well as the total predicted PKA phosphorylation sites for each mutant (mean of three independent experiments ± SD). (F) Native gel electrophoresis indicates that destabilization of pentameric Npm-N by the double T95D/S125E mutations, mimicking solvent-exposed phosphorylation, allows PKA phosphorylation, which consequently drives dissociation of the pentamer into monomers (red arrow).
Fig. 4.
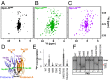
Peptides containing and R-rich motif bind to Npm-N pentamer and promote assembly of Npm-N monomers. (A) Titrations of ARF-N37 (0, 1.0, 2.5, 5.0, 7.5, and 10 μM) in WT and S88E (10 μM) assayed by native gel electrophoresis. (B) Histograms of chemical shift perturbations observed in 2D TROSY correlation spectra of 0.4 mM [15N]Npm-N pentamer upon interaction with 3× molar excess of Arf6 (green), Rev37-47 (purple), and rpL521-37 (blue) peptides. (C) Arg side chains of Arf6 peptide form electrostatic contacts with acidic groups from adjacent protomers within Npm-N pentamer; one molecule of Arf6 (blue) is shown docked between chains A (light blue) and E (light magenta) shown in surface representation, with the residues perturbed by Arf6 interaction highlighted in red.
Fig. 5.
Navigating the Npm-N conformational landscape. Structural polymorphism of Npm-N is facilitated by repulsive electrostatic forces as a result of close spatial proximity of acidic tracts A1 and A2 (red lines) within the pentameric structure. Documented and putative solvent-exposed and buried phosphorylation sites are illustrated (green dots). Unphosphorylated Npm-N predominantly populates the pentameric state (Right). Phosphorylation (red circles) of pentameric Npm-N at solvent exposed sites slightly destabilizes the oligomeric structure (indicated by wavy yellow lines), reducing the energy barrier to make the conformational transition to other, less native-like structures, thus exposing additional, otherwise buried sites for subsequent phosphorylation. Phosphorylation at buried sites dramatically destabilizes the oligomeric structure, stabilizing monomeric folded or monomeric disordered structures (Left). Both destabilization scenarios will expose additional interior sites for multisite phosphorylation to structurally lock Npm-N in the monomeric, disordered state. This process can be reversed by dephosphorylation by phosphatases. In addition, binding to target proteins containing R-rich motifs stabilizes the pentameric form, counteracting the destabilization associated with phosphorylation.
Similar articles
- Dynamic conformations of nucleophosmin (NPM1) at a key monomer-monomer interface affect oligomer stability and interactions with granzyme B.
Duan-Porter WD, Woods VL Jr, Maurer KD, Li S, Rosen A. Duan-Porter WD, et al. PLoS One. 2014 Dec 9;9(12):e115062. doi: 10.1371/journal.pone.0115062. eCollection 2014. PLoS One. 2014. PMID: 25490769 Free PMC article. - Structural basis for the recognition by 14-3-3 proteins of a conditional binding site within the oligomerization domain of human nucleophosmin.
Kapitonova AA, Tugaeva KV, Varfolomeeva LA, Boyko KM, Cooley RB, Sluchanko NN. Kapitonova AA, et al. Biochem Biophys Res Commun. 2022 Oct 30;627:176-183. doi: 10.1016/j.bbrc.2022.08.047. Epub 2022 Aug 23. Biochem Biophys Res Commun. 2022. PMID: 36041327 - Cryptic disorder: an order-disorder transformation regulates the function of nucleophosmin.
Mitrea DM, Kriwacki RW. Mitrea DM, et al. Pac Symp Biocomput. 2012:152-63. Pac Symp Biocomput. 2012. PMID: 22174271 - Nucleophosmin, a multifunctional nucleolar organizer with a role in DNA repair.
López DJ, Rodríguez JA, Bañuelos S. López DJ, et al. Biochim Biophys Acta Proteins Proteom. 2020 Dec;1868(12):140532. doi: 10.1016/j.bbapap.2020.140532. Epub 2020 Aug 25. Biochim Biophys Acta Proteins Proteom. 2020. PMID: 32853771 Review. - Translocations and mutations involving the nucleophosmin (NPM1) gene in lymphomas and leukemias.
Falini B, Nicoletti I, Bolli N, Martelli MP, Liso A, Gorello P, Mandelli F, Mecucci C, Martelli MF. Falini B, et al. Haematologica. 2007 Apr;92(4):519-32. doi: 10.3324/haematol.11007. Haematologica. 2007. PMID: 17488663 Review.
Cited by
- LASSI: A lattice model for simulating phase transitions of multivalent proteins.
Choi JM, Dar F, Pappu RV. Choi JM, et al. PLoS Comput Biol. 2019 Oct 21;15(10):e1007028. doi: 10.1371/journal.pcbi.1007028. eCollection 2019 Oct. PLoS Comput Biol. 2019. PMID: 31634364 Free PMC article. - Liquid-liquid phase separation in tumor biology.
Tong X, Tang R, Xu J, Wang W, Zhao Y, Yu X, Shi S. Tong X, et al. Signal Transduct Target Ther. 2022 Jul 8;7(1):221. doi: 10.1038/s41392-022-01076-x. Signal Transduct Target Ther. 2022. PMID: 35803926 Free PMC article. Review. - What's in an Average? An Ensemble View of Phosphorylation Effects.
Chin AF, Hilser VJ. Chin AF, et al. Structure. 2017 Apr 4;25(4):573-575. doi: 10.1016/j.str.2017.03.012. Structure. 2017. PMID: 28380337 Free PMC article. - Localization of AML-related nucleophosmin mutant depends on its subtype and is highly affected by its interaction with wild-type NPM.
Brodská B, Kráčmarová M, Holoubek A, Kuželová K. Brodská B, et al. PLoS One. 2017 Apr 6;12(4):e0175175. doi: 10.1371/journal.pone.0175175. eCollection 2017. PLoS One. 2017. PMID: 28384310 Free PMC article. - Liquid-liquid phase separation in DNA double-strand breaks repair.
Wang YL, Zhao WW, Shi J, Wan XB, Zheng J, Fan XJ. Wang YL, et al. Cell Death Dis. 2023 Nov 15;14(11):746. doi: 10.1038/s41419-023-06267-0. Cell Death Dis. 2023. PMID: 37968256 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM083159/GM/NIGMS NIH HHS/United States
- P30CA21765/CA/NCI NIH HHS/United States
- P30 CA021765/CA/NCI NIH HHS/United States
- 5R01-CAO82491/PHS HHS/United States
- 5R01-GM083159/GM/NIGMS NIH HHS/United States
- R25 CA023944/CA/NCI NIH HHS/United States
- MC_U105185859/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases