Detection of viral proteins in human cells lines by xeno-proteomics: elimination of the last valid excuse for not testing every cellular proteome dataset for viral proteins - PubMed (original) (raw)
Detection of viral proteins in human cells lines by xeno-proteomics: elimination of the last valid excuse for not testing every cellular proteome dataset for viral proteins
Alexey L Chernobrovkin et al. PLoS One. 2014.
Abstract
Cell cultures used routinely in proteomic experiments may contain proteins from other species because of infection, transfection or just contamination. Since infection or contamination may affect the results of a biological experiment, it is important to test the samples for the presence of "alien" proteins. Usually cells are tested only for the most common infections, and most of the existing tests are targeting specific contaminations. Here we describe a three-step procedure for reliable untargeted detection of viral proteins using proteomics data, and recommend this or similar procedure to be applied to every proteomics dataset submitted for publication.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
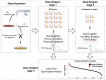
Figure 1. The suggested three-step workflow of shotgun proteomics analysis that includes contamination/infection detection.
(1) conventional matching the experimental MS/MS spectra against the protein database of the host organism (human); (2) Unmatched MS/MS spectra and insignificant matches in terms of FDR are matched against viral protein database. (3) The presence of significant hits at the second step points to the potential infection/contamination of the sample. Label-free quantitative analysis of identified proteins (both host and contaminant) provides an estimation of the contamination level and thus severity of the problem.
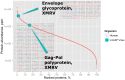
Figure 2. Expression levels of the XMRV viral proteins within the proteome of the human cell line LNCaP.
Small red dots correspond to the host cell proteins, whereas the two identified viral proteins are marked as cyan circles. Detected sequences of tryptic peptides of the two viral proteins are marked with color: red - peptides mapped perfectly on the XMRV sequence; blue and green - mutated sequences.
Similar articles
- Moloney murine leukemia virus glyco-gag facilitates xenotropic murine leukemia virus-related virus replication through human APOBEC3-independent mechanisms.
Nitta T, Lee S, Ha D, Arias M, Kozak CA, Fan H. Nitta T, et al. Retrovirology. 2012 Jul 24;9:58. doi: 10.1186/1742-4690-9-58. Retrovirology. 2012. PMID: 22828015 Free PMC article. - Investigation of xenotropic murine leukemia virus-related virus (XMRV) in human and other cell lines.
Williams DK, Galvin TA, Ma H, Khan AS. Williams DK, et al. Biologicals. 2011 Nov;39(6):378-83. doi: 10.1016/j.biologicals.2011.08.011. Epub 2011 Oct 12. Biologicals. 2011. PMID: 21996050 - Time-resolved Global and Chromatin Proteomics during Herpes Simplex Virus Type 1 (HSV-1) Infection.
Kulej K, Avgousti DC, Sidoli S, Herrmann C, Della Fera AN, Kim ET, Garcia BA, Weitzman MD. Kulej K, et al. Mol Cell Proteomics. 2017 Apr;16(4 suppl 1):S92-S107. doi: 10.1074/mcp.M116.065987. Epub 2017 Feb 8. Mol Cell Proteomics. 2017. PMID: 28179408 Free PMC article. - Viral proteomics.
Maxwell KL, Frappier L. Maxwell KL, et al. Microbiol Mol Biol Rev. 2007 Jun;71(2):398-411. doi: 10.1128/MMBR.00042-06. Microbiol Mol Biol Rev. 2007. PMID: 17554050 Free PMC article. Review. - Investigating the biology of alpha herpesviruses with MS-based proteomics.
Engel EA, Song R, Koyuncu OO, Enquist LW. Engel EA, et al. Proteomics. 2015 Jun;15(12):1943-56. doi: 10.1002/pmic.201400604. Epub 2015 May 15. Proteomics. 2015. PMID: 25764121 Free PMC article. Review.
Cited by
- SpotLight Proteomics: uncovering the hidden blood proteome improves diagnostic power of proteomics.
Lundström SL, Zhang B, Rutishauser D, Aarsland D, Zubarev RA. Lundström SL, et al. Sci Rep. 2017 Feb 7;7:41929. doi: 10.1038/srep41929. Sci Rep. 2017. PMID: 28167817 Free PMC article. - Tiered Human Integrated Sequence Search Databases for Shotgun Proteomics.
Deutsch EW, Sun Z, Campbell DS, Binz PA, Farrah T, Shteynberg D, Mendoza L, Omenn GS, Moritz RL. Deutsch EW, et al. J Proteome Res. 2016 Nov 4;15(11):4091-4100. doi: 10.1021/acs.jproteome.6b00445. Epub 2016 Sep 12. J Proteome Res. 2016. PMID: 27577934 Free PMC article.
References
- Gillet J-P, Varma S, Gottesman MM (2013) The Clinical Relevance of Cancer Cell Lines. J Natl Cancer Inst: djt007–. Available: http://jnci.oxfordjournals.org/content/early/2013/02/21/jnci.djt007.short. Accessed 2013 March 4. - PMC - PubMed
- Garnett MJ, Edelman EJ, Heidorn SJ, Greenman CD, Dastur A, et al.. (2012) Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature 483: 570–575. Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3349233&tool=p.... Accessed 2013 Feb 27. - PMC - PubMed
- Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin A a, et al.. (2012) The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483: 603–607. Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3320027&tool=p.... Accessed 2013 Feb 28. - PMC - PubMed
- Mobley A, Linder SK, Braeuer R, Ellis LM, Zwelling L (2013) A survey on data reproducibility in cancer research provides insights into our limited ability to translate findings from the laboratory to the clinic. PLoS One 8: e63221. Available: http://dx.plos.org/10.1371/journal.pone.0063221. Accessed 2013 Aug 2. - DOI - PMC - PubMed
- Drexler HG, Uphoff CC (2002) Mycoplasma contamination of cell cultures: Incidence, sources, effects, detection, elimination, prevention. Cytotechnology 39: 75–90 Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3463982&tool=p.... - PMC - PubMed
MeSH terms
Substances
Grants and funding
The authors have no support or funding to report.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials