The treatment-naive microbiome in new-onset Crohn's disease - PubMed (original) (raw)
. 2014 Mar 12;15(3):382-392.
doi: 10.1016/j.chom.2014.02.005.
Subra Kugathasan # 2, Lee A Denson # 3, Yoshiki Vázquez-Baeza 4, Will Van Treuren 5, Boyu Ren 6, Emma Schwager 6, Dan Knights 7 8, Se Jin Song 5, Moran Yassour 1, Xochitl C Morgan 6, Aleksandar D Kostic 1, Chengwei Luo 1, Antonio González 5, Daniel McDonald 5, Yael Haberman 3, Thomas Walters 9, Susan Baker 10, Joel Rosh 11, Michael Stephens 12, Melvin Heyman 13, James Markowitz 14, Robert Baldassano 15, Anne Griffiths 16, Francisco Sylvester 17, David Mack 18, Sandra Kim 19, Wallace Crandall 19, Jeffrey Hyams 17, Curtis Huttenhower 1 6, Rob Knight 5 20 21, Ramnik J Xavier 1 22 23
Affiliations
- PMID: 24629344
- PMCID: PMC4059512
- DOI: 10.1016/j.chom.2014.02.005
The treatment-naive microbiome in new-onset Crohn's disease
Dirk Gevers et al. Cell Host Microbe. 2014.
Abstract
Inflammatory bowel diseases (IBDs), including Crohn's disease (CD), are genetically linked to host pathways that implicate an underlying role for aberrant immune responses to intestinal microbiota. However, patterns of gut microbiome dysbiosis in IBD patients are inconsistent among published studies. Using samples from multiple gastrointestinal locations collected prior to treatment in new-onset cases, we studied the microbiome in the largest pediatric CD cohort to date. An axis defined by an increased abundance in bacteria which include Enterobacteriaceae, Pasteurellacaea, Veillonellaceae, and Fusobacteriaceae, and decreased abundance in Erysipelotrichales, Bacteroidales, and Clostridiales, correlates strongly with disease status. Microbiome comparison between CD patients with and without antibiotic exposure indicates that antibiotic use amplifies the microbial dysbiosis associated with CD. Comparing the microbial signatures between the ileum, the rectum, and fecal samples indicates that at this early stage of disease, assessing the rectal mucosal-associated microbiome offers unique potential for convenient and early diagnosis of CD.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures
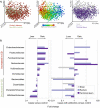
Figure 1. Most differential taxa in pediatric CD
(A) A set of principal coordinate plots of the unweighted UniFrac distance, with each sample colored either by the disease phenotype (left), alpha diversity (middle), or sample type (right). PC1, PC2, and PC3 represent the top three principal coordinates that captured most of the diversity, with the fraction of diversity captured by that coordinate shown in percent. (B) Differences in abundance are shown for the taxonomic biomarkers that were detected using a multivariate statistical approach (see Experimental Procedures and Table S2). The fold change for each taxon was calculated by dividing the mean abundance in the cases by that of the controls. Several taxonomic biomarkers measured at both the ileal and rectal sites were found to be significantly correlated with disease phenotype; however, most of that microbial signal was lost in the stool samples. The fraction of patients that were on antibiotics during sample collection was considered as an individual subtype, due to the large confounding impact antibiotic exposure causes on the microbial composition (see Table S2). The left shows cases without antibiotic treatment, and the right includes the fraction of cases (10%) that were under antibiotic pressure at sampling. The taxa at the top are increased in disease state, whereas the taxa at the bottom follow an opposite trend. Apparent missing bars are cases in which there is no difference, or fold change equals 1. Use of antibiotics does impact the microbial composition by tipping the microbial community further towards a dysbiotic state, and has a differential impact on the taxa, depending on organism and sampling site. Related to Figure S1, Table S1.
Figure 2. The Microbial Dysbiosis index characterizes CD severity
(A) A correlation network was inferred for the ileal microbiota compositions using CCREPE with a checkerboard score, indicating a strong co-occurrence between taxa of the same disease-associated behavior and a co-exclusion between taxa of a different behavior. Nodes represent the different taxa, and color corresponds to their behavior in disease, with green for those decreased in CD and red for those increased in CD. Edges between nodes represent correlations between the nodes they connect, with edge colors of dark and light grey indicating positive and negative correlations, respectively. For clarity, only edges corresponding to correlations whose similarity was less than 0.3 are shown. (B) Scatterplot of the arcsine square root transformed abundances of all summed abundances for the taxa increased (top) or decreased (bottom) in CD, versus the pediatric CD activity index (PCDAI (Hyams et al., 1991)) as a measure for disease severity. (C) Scatter density plot of the species richness (Chao1, (Chao et al., 2006)) versus the Microbial Dysbiosis index (MD-index) for each sample. The increase in blue color (white to dark blue) reflects the density of the scatter plot. The MD-index is defined as the log of [total abundance in organisms increased in CD] over [total abundance of organisms decreased in CD] (organisms listed in Fig 1A), and is intended as an overall summary statistic for the microbial dysbiosis described in more detail in panel A. In samples with a high MD-index (> 1), a strong reduction in the species richness was observed. (D) A principal coordinate plot of the unweighted UniFrac distance, colored by the MD-index. Sqrt, square root.Related to Figure S2, Table S2.
Figure 3. Comparative genomics of CD biomarkers
The KEGG metabolic pathways that differentiate the species by behavior in disease state are shown as a heatmap. A selection of reference genomes that are representative for the species increased or decreased with disease were obtained from IMG (JGI), and biomarker detection was performed on their gene content at the level of KEGG pathways. Several were statistically significant (Wilcoxon, p < 10e-8) and are visualized here. Related to Table S2.
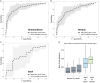
Figure 4. Disease classification performs well on biopsy-associated microbiome profiles
(A-C) For each of the three sample types, including terminal ileum biopsy (A), rectum biopsy (B), and stool sample (C), we evaluated the accuracy of disease classification using L1 penalized logistic regression with ROC curves representing the results. Dashed lines show the mean performance obtained when genus-level features were used, and the surrounding grey area is the 95% confidence interval. Terminal ileum biopsies performed best (AUC = 0.85), closely matched by the rectum biopsies (AUC = 0.78). However, the classifier based on the stool samples collected at the time of the diagnosis performs less well (AUC = 0.66), and with low consistency (large confidence interval). (D) The intra-subject diversity in microbiome composition was determined for all pairwise sample type combinations. Both biopsy samples were found to be highly similar, whereas the stool sample was quite diverse. Further, we also compared whether disease location would impact the intra-subject diversity between the two tissue biopsy locations. The location of the disease, ileal (L1), colonic (L2), or ileocolonic (L3), did not significantly disrupt the similarity between the two intra-subject mucosal-associated microbiota. Also, no biomarker was detected allowing us to distinguish these disease sub-phenotypes. Related to Figure S3.
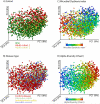
Figure 5. A view of the microbial composition across different IBD cohorts
We combined microbial profiles obtained for 1,742 subjects from three different IBD cohorts and generated a set of principal coordinate plots of the unweighted UniFrac distance, where each sample was colored by (A) cohort, (B) disease type, (C) MD-index, or (D) species richness (Chao1). From this combined view, it is clear that the first principal coordinate (PC1) stratifies the samples by species richness, which is negatively correlated with MD-index, and that the second principal coordinate (PC2) is largely affected by cohort. Disease phenotype is no obvious driver for sample clustering.
Comment in
- Microbiome: bacterial imbalance in Crohn's disease.
Hofer U. Hofer U. Nat Rev Microbiol. 2014 May;12(5):312. doi: 10.1038/nrmicro3255. Epub 2014 Mar 17. Nat Rev Microbiol. 2014. PMID: 24638106 No abstract available. - IBD. Understanding gut microbiota in new-onset Crohn's disease.
Ray K. Ray K. Nat Rev Gastroenterol Hepatol. 2014 May;11(5):268. doi: 10.1038/nrgastro.2014.45. Epub 2014 Mar 25. Nat Rev Gastroenterol Hepatol. 2014. PMID: 24662277 No abstract available. - Gut microbiome in new-onset Crohn's disease.
Hall LJ, Walshaw J, Watson AJ. Hall LJ, et al. Gastroenterology. 2014 Oct;147(4):932-4. doi: 10.1053/j.gastro.2014.08.014. Epub 2014 Aug 23. Gastroenterology. 2014. PMID: 25152198 No abstract available.
Similar articles
- Dysbiosis, inflammation, and response to treatment: a longitudinal study of pediatric subjects with newly diagnosed inflammatory bowel disease.
Shaw KA, Bertha M, Hofmekler T, Chopra P, Vatanen T, Srivatsa A, Prince J, Kumar A, Sauer C, Zwick ME, Satten GA, Kostic AD, Mulle JG, Xavier RJ, Kugathasan S. Shaw KA, et al. Genome Med. 2016 Jul 13;8(1):75. doi: 10.1186/s13073-016-0331-y. Genome Med. 2016. PMID: 27412252 Free PMC article. - Gut microbiome in new-onset Crohn's disease.
Hall LJ, Walshaw J, Watson AJ. Hall LJ, et al. Gastroenterology. 2014 Oct;147(4):932-4. doi: 10.1053/j.gastro.2014.08.014. Epub 2014 Aug 23. Gastroenterology. 2014. PMID: 25152198 No abstract available. - IBD. Understanding gut microbiota in new-onset Crohn's disease.
Ray K. Ray K. Nat Rev Gastroenterol Hepatol. 2014 May;11(5):268. doi: 10.1038/nrgastro.2014.45. Epub 2014 Mar 25. Nat Rev Gastroenterol Hepatol. 2014. PMID: 24662277 No abstract available. - Gut dysbiosis and paediatric Crohn's disease.
Brusaferro A, Cavalli E, Farinelli E, Cozzali R, Principi N, Esposito S. Brusaferro A, et al. J Infect. 2019 Jan;78(1):1-7. doi: 10.1016/j.jinf.2018.10.005. Epub 2018 Oct 16. J Infect. 2019. PMID: 30336176 Review. - The Microbiome in Crohn's Disease: Role in Pathogenesis and Role of Microbiome Replacement Therapies.
Khanna S, Raffals LE. Khanna S, et al. Gastroenterol Clin North Am. 2017 Sep;46(3):481-492. doi: 10.1016/j.gtc.2017.05.004. Epub 2017 Jul 19. Gastroenterol Clin North Am. 2017. PMID: 28838410 Review.
Cited by
- Metabolic modelling reveals broad changes in gut microbial metabolism in inflammatory bowel disease patients with dysbiosis.
Heinken A, Hertel J, Thiele I. Heinken A, et al. NPJ Syst Biol Appl. 2021 May 6;7(1):19. doi: 10.1038/s41540-021-00178-6. NPJ Syst Biol Appl. 2021. PMID: 33958598 Free PMC article. - Fecal microbiome in systemic sclerosis, in search for the best candidate for microbiota-targeted therapy for small intestinal bacterial overgrowth control.
Fiorentini E, Russo E, Amedei A, Bellando Randone S. Fiorentini E, et al. J Scleroderma Relat Disord. 2022 Oct;7(3):163-167. doi: 10.1177/23971983221118871. Epub 2022 Sep 25. J Scleroderma Relat Disord. 2022. PMID: 36211209 Free PMC article. - The effect of NOD2 on the microbiota in Crohn's disease.
Lauro ML, Burch JM, Grimes CL. Lauro ML, et al. Curr Opin Biotechnol. 2016 Aug;40:97-102. doi: 10.1016/j.copbio.2016.02.028. Epub 2016 Mar 29. Curr Opin Biotechnol. 2016. PMID: 27035071 Free PMC article. Review. - Center Variation in Intestinal Microbiota Prior to Late-Onset Sepsis in Preterm Infants.
Taft DH, Ambalavanan N, Schibler KR, Yu Z, Newburg DS, Deshmukh H, Ward DV, Morrow AL. Taft DH, et al. PLoS One. 2015 Jun 25;10(6):e0130604. doi: 10.1371/journal.pone.0130604. eCollection 2015. PLoS One. 2015. PMID: 26110908 Free PMC article. - Increased Intestinal Microbial Diversity Following Fecal Microbiota Transplant for Active Crohn's Disease.
Vaughn BP, Vatanen T, Allegretti JR, Bai A, Xavier RJ, Korzenik J, Gevers D, Ting A, Robson SC, Moss AC. Vaughn BP, et al. Inflamm Bowel Dis. 2016 Sep;22(9):2182-90. doi: 10.1097/MIB.0000000000000893. Inflamm Bowel Dis. 2016. PMID: 27542133 Free PMC article.
References
- Chao A, Chazdon RL, Colwell RK, Shen TJ. Abundance-based similarity indices and their estimation when there are unseen species in samples. Biometrics. 2006;62:361–371. - PubMed
- Chavan RS, Pannaraj PS, Luna RA, Szabo S, Adesina A, Versalovic J, Krance RA, Kennedy-Nasser AA. Significant morbidity and mortality attributable to rothia mucilaginosa infections in children with hematological malignancies or following hematopoietic stem cell transplantation. Pediatr Hematol Oncol. 2013;30:445–454. - PubMed
Publication types
MeSH terms
Grants and funding
- P30 DK043351/DK/NIDDK NIH HHS/United States
- R01 DK092405/DK/NIDDK NIH HHS/United States
- R01DK092405/DK/NIDDK NIH HHS/United States
- T32 GM074897/GM/NIGMS NIH HHS/United States
- U54 DE023798/DE/NIDCR NIH HHS/United States
- R01 HG005969/HG/NHGRI NIH HHS/United States
- U54 DK102557/DK/NIDDK NIH HHS/United States
- R35 CA197449/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases