Spliced X-box binding protein 1 couples the unfolded protein response to hexosamine biosynthetic pathway - PubMed (original) (raw)
. 2014 Mar 13;156(6):1179-1192.
doi: 10.1016/j.cell.2014.01.014.
Yingfeng Deng 2, Ningguo Gao 3, Zully Pedrozo 4, Dan L Li 1, Cyndi R Morales 1, Alfredo Criollo 5, Xiang Luo 1, Wei Tan 1, Nan Jiang 1, Mark A Lehrman 3, Beverly A Rothermel 6, Ann-Hwee Lee 7, Sergio Lavandero 4, Pradeep P A Mammen 1, Anwarul Ferdous 1, Thomas G Gillette 1, Philipp E Scherer 8, Joseph A Hill 9
Affiliations
- PMID: 24630721
- PMCID: PMC3959665
- DOI: 10.1016/j.cell.2014.01.014
Spliced X-box binding protein 1 couples the unfolded protein response to hexosamine biosynthetic pathway
Zhao V Wang et al. Cell. 2014.
Abstract
The hexosamine biosynthetic pathway (HBP) generates uridine diphosphate N-acetylglucosamine (UDP-GlcNAc) for glycan synthesis and O-linked GlcNAc (O-GlcNAc) protein modifications. Despite the established role of the HBP in metabolism and multiple diseases, regulation of the HBP remains largely undefined. Here, we show that spliced X-box binding protein 1 (Xbp1s), the most conserved signal transducer of the unfolded protein response (UPR), is a direct transcriptional activator of the HBP. We demonstrate that the UPR triggers HBP activation via Xbp1s-dependent transcription of genes coding for key, rate-limiting enzymes. We further establish that this previously unrecognized UPR-HBP axis is triggered in a variety of stress conditions. Finally, we demonstrate a physiologic role for the UPR-HBP axis by showing that acute stimulation of Xbp1s in heart by ischemia/reperfusion confers robust cardioprotection in part through induction of the HBP. Collectively, these studies reveal that Xbp1s couples the UPR to the HBP to protect cells under stress.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures
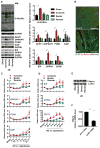
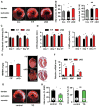
Figure 1. Induction of HBP, O-GlcNAc protein modification, and the UPR in heart by ischemia/reperfusion (I/R)
(A) Protein O-GlcNAc modification was increased in the infarct zone of I/R-stressed heart. Protein levels of the hexosamine biosynthetic pathway (HBP) and UPR markers were elevated in the same region. GAPDH was used as loading control. N = 3 for each group. (B) Protein expression and localization of GFAT1 were visualized by fluorescence immunostaining in heart tissue from sham-operated and I/R animals. Arrows indicate cardiomyocytes in the ischemic region. Scale bars: 50 μm. (C) Transcription of the HBP genes, GFAT1, GNPNAT1, and PGM3, as well as GalE, was induced in the infarct region of heart during I/R as assessed by qRT-PCR. Sham samples of 4 hrs and 24 hrs were pooled and used as control. N = 3–9. (D) Transcription of UPR genes was induced in the infarct region of heart during I/R as assessed by qRT-PCR. N = 3–9. (E) Xbp1s was increased in the infarct region of I/R-stressed heart. Lamin was used as loading control for nuclear extracts. * denotes a non-specific signal across all samples. (F) Xbp1s mRNA levels were reduced in human heart following LVAD mechanical support. N = 8. Data are represented as mean ± SEM. *, p < 0.05, **, p < 0.01. See also Figure S1, Table S2.
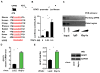
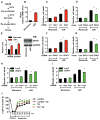
Figure 2. GFAT1 is a direct target of Xbp1s
(A) A conserved DNA motif, similar to the UPRE, was identified in the GFAT1 promoter from different species. (B) GFAT1 promoter was stimulated by Xbp1s overexpression. The GFAT1 promoter activity was measured by a luciferase assay upon Xbp1s co-transfection in HEK293T cells. N = 3 for each group. (C) Xbp1s was associated with A ChIP assay was conducted in C2C12 cells after Xbp1s overexpression. PCR amplification was performed using primers spanning the UPRE or from a distal region in the GFAT1 promoter. The triangles indicate increasing amounts of immunoprecipitated DNA for PCR reaction. (D) GFAT1 transcription was stimulated by Xbp1s in vitro. qPCR was conducted to quantify relative mRNA levels of GFAT1 after Xbp1s overexpression in NRVM. N = 6. (E) GFAT1 protein levels were elevated by Xbp1s induction. N = 3–4. Data are represented as mean ± SEM. *, p < 0.05. See also Figure S2.
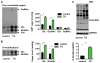
Figure 3. Xbp1s drives GFAT1 expression in vivo
(A) Inducible overexpression of Xbp1s in cardiomyocytes in vivo. Xbp1s expression was suppressed by doxycycline (Dox). (B) GFAT1 transcription was significantly induced by Xbp1s overexpression. Control (αMHC-tTA only) and TG (αMHC-tTA, TRE-Xbp1s double transgenic) mice were placed on regular water (1–3 weeks) to stimulate Xbp1s expression. Cardiac GFAT1 mRNA levels were quantified by qRT-PCR. N = 3. (C) GFAT1 protein expression was compared between control and TG mouse hearts after 2-week induction of Xbp1s. GAPDH was used as loading control. N = 3. Data are represented as mean ± SEM. *, p < 0.05. (D) GFAT1 protein levels and expression patterns were assessed by immunostaining. Scale bars: 50 μm. See also Figure S4.
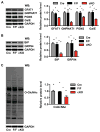
Figure 4. Xbp1s induction leads to increases in nucleotide sugars and O-GlcNAc modification
(A) Induction of Xbp1s in cardiomyocytes led to significant increases in free nucleotide sugars in heart. Control and TG mice were placed on regular water (2 weeks) to stimulate Xbp1s expression. Free nucleotide sugars were analyzed by FACE. Samples corresponding to equal amount of total cellular proteins were loaded for each mouse strain. N = 6. (B) Induction of Xbp1s in cardiomyocytes led to augmentation of O-Glc and O-GlcNAc protein modifications. Total cellular O-linked monosaccharides were separated by FACE gel. N = 6 for each group. (C) Cardiac induction of Xbp1s led to significant increases in O-GlcNAc protein modification, as evaluated by immunoblotting for O-GlcNAc. GAPDH was used as loading control. N = 6 for control and n = 8 for TG mice. Data are represented as mean ± SEM. *, p < 0.05, **, p < 0.01. See also Figure S4.
Figure 5. Xbp1s is required for induction of the UPR, the HBP, and O-GlcNAc modification in heart after I/R
(A) Induction of the HBP genes, GFAT1, GNPNAT1, and PGM3, as well as GalE was analyzed in αMHC-Cre, F/F and cKO hearts 24 hrs after I/R. N = 3–4 per group. (B) Expression of the UPR markers BiP and GRP94 was analyzed 24 hrs after I/R. N = 3–4. (C) O-GlcNAc modification was reduced in cKO hearts compared with controls. N = 3–4. Data are represented as mean ± SEM. *, p < 0.05. See also Figure S3, S5.
Figure 6. Xbp1s induction protects heart from I/R injury in vivo
(A) Xbp1 silencing led to increased injury from I/R. Male mice were subjected I/R. Cardiac injury was assayed by TTC staining. Blue, unaffected, viable tissue; red, area at risk; white, infarct area. Scale bars: 1 mm. (B) Infarct area (relative to area at risk) and area at risk (relative to left ventricle) were quantified. Number of animals used is indicated. (C) Ventricular function in cKO mice manifested significant deterioration as measured by % fractional shortening 7 days and 21 days post I/R. N = 5 for F/F and n = 7 for cKO. (D) Left ventricular internal diameters (LVID) in diastole (left) and systole (right) were compared. N = 5–7. (E) cKO mice developed more severe hypertrophy following I/R. N = 3–5. Scale bars: 2 mm. (F) cKO mice manifested more robust fetal gene reactivation after I/R. N = 3–5. (G) Xbp1s overexpression protected hearts from I/R injury. Control and TG mice were placed on normal water for 2 weeks to induce transgene expression. I/R surgery was performed and cardiac injury was assessed by TTC staining. Scale bars: 1 mm. (H) Infarct area (relative to area at risk) and area at risk (relative to left ventricle) were quantified. Data are represented as mean ± SEM. *, p < 0.05, **, p < 0.01. NS, not significant. See also Figure S5, S6.
Figure 7. GFAT1 is required for Xbp1s-dependent cardioprotection during I/R
(A) Experimental procedures for NRVM in vitro. (B) Simulated I/R (sI/R) induced Xbp1s expression as assessed by qRT-PCR. N = 3. (C) Knockdown of Xbp1 led to enhanced cell death in response to sI/R. Xbp1s expression was reduced by siRNA. Cell death was measured by LDH release. N = 3 for each group. (D) Xbp1s overexpression protected cardiomyocytes from sI/R injury. NRVM were infected with lentivirus expressing either LacZ or Xbp1s. Cell death from sI/R was quantified by LDH measurements. N = 6. (E) GFAT1 mRNA and protein levels were significantly induced in NRVM after sI/R. N = 3. (F) Knockdown of GFAT1 exacerbated sI/R injury. GFAT1 expression was reduced by siRNA. NRVM were then subjected to sI/R, and cell death was assessed by LDH release. N = 3. (G) GFAT1 overexpression protected cardiomyocytes from sI/R injury. NRVM were infected with lentivirus expressing either LacZ or GFAT1. Cell death from sI/R was quantified by LDH measurements. N = 6 for each group. (H) GFAT1 knockdown diminished Xbp1s cardioprotection in sI/R. NRVM were first infected with LacZ or Xbp1s lentivirus. GFAT1 was reduced by siRNA. After sI/R, LDH assays were conducted. N = 3. (I) Overexpression of GFAT1 significantly rescued cell death by Xbp1 silencing. NRVM were first infected with LacZ or GFAT1 lentivirus. Xbp1 was reduced by siRNA. After sI/R, cell death was quantified by LDH assay. N = 6 for each group. Data are represented as mean ± SEM. *, p < 0.05, **, p < 0.01. (J) Inhibition of GFAT1 diminished cardioprotection by Xbp1s. Control and TG mouse hearts were processed for Langendorff analysis (ischemia 20 min; reperfusion 40 min). Cardiac function was assessed as left ventricular developed pressure (LVDP). N = 7 for control, n = 4 for TG, n = 4 for control + Azaserine (Az), and n = 4 for TG + Az. Data are represented as mean ± SEM. *, TG vs control, p < 0.05. #, TG vs TG + Az, p < 0.05. See also Figure S7.
Comment in
- Sugarcoating ER Stress.
Vincenz L, Hartl FU. Vincenz L, et al. Cell. 2014 Mar 13;156(6):1125-1127. doi: 10.1016/j.cell.2014.02.035. Cell. 2014. PMID: 24630714 - Finding the missing link between the unfolded protein response and O-GlcNAcylation in the heart.
Glembotski CC. Glembotski CC. Circ Res. 2014 Aug 29;115(6):546-8. doi: 10.1161/CIRCRESAHA.114.304855. Circ Res. 2014. PMID: 25170091 Free PMC article.
Similar articles
- Sugarcoating ER Stress.
Vincenz L, Hartl FU. Vincenz L, et al. Cell. 2014 Mar 13;156(6):1125-1127. doi: 10.1016/j.cell.2014.02.035. Cell. 2014. PMID: 24630714 - Paramyxovirus replication induces the hexosamine biosynthetic pathway and mesenchymal transition via the IRE1α-XBP1s arm of the unfolded protein response.
Qiao D, Skibba M, Xu X, Garofalo RP, Zhao Y, Brasier AR. Qiao D, et al. Am J Physiol Lung Cell Mol Physiol. 2021 Sep 1;321(3):L576-L594. doi: 10.1152/ajplung.00127.2021. Epub 2021 Jul 28. Am J Physiol Lung Cell Mol Physiol. 2021. PMID: 34318710 Free PMC article. - Tisp40 prevents cardiac ischemia/reperfusion injury through the hexosamine biosynthetic pathway in male mice.
Zhang X, Hu C, Ma ZG, Hu M, Yuan XP, Yuan YP, Wang SS, Kong CY, Teng T, Tang QZ. Zhang X, et al. Nat Commun. 2023 Jun 8;14(1):3383. doi: 10.1038/s41467-023-39159-0. Nat Commun. 2023. PMID: 37291168 Free PMC article. - Hexosamines, insulin resistance, and the complications of diabetes: current status.
Buse MG. Buse MG. Am J Physiol Endocrinol Metab. 2006 Jan;290(1):E1-E8. doi: 10.1152/ajpendo.00329.2005. Am J Physiol Endocrinol Metab. 2006. PMID: 16339923 Free PMC article. Review. - Dysregulation of hexosamine biosynthetic pathway wiring metabolic signaling circuits in cancer.
Itano N, Iwamoto S. Itano N, et al. Biochim Biophys Acta Gen Subj. 2023 Jan;1867(1):130250. doi: 10.1016/j.bbagen.2022.130250. Epub 2022 Oct 10. Biochim Biophys Acta Gen Subj. 2023. PMID: 36228878 Review.
Cited by
- Beyond adiponectin and leptin: adipose tissue-derived mediators of inter-organ communication.
Funcke JB, Scherer PE. Funcke JB, et al. J Lipid Res. 2019 Oct;60(10):1648-1684. doi: 10.1194/jlr.R094060. Epub 2019 Jun 17. J Lipid Res. 2019. PMID: 31209153 Free PMC article. Review. - Best practices in assessing cardiac protein _O_-GlcNAcylation by immunoblot.
Zou L, Zhang D, Ha CM, Wende AR, Chatham JC. Zou L, et al. Am J Physiol Heart Circ Physiol. 2023 Oct 1;325(4):H601-H616. doi: 10.1152/ajpheart.00104.2023. Epub 2023 Aug 4. Am J Physiol Heart Circ Physiol. 2023. PMID: 37539459 Free PMC article. - Mesencephalic astrocyte-derived neurotrophic factor is an ER-resident chaperone that protects against reductive stress in the heart.
Arrieta A, Blackwood EA, Stauffer WT, Santo Domingo M, Bilal AS, Thuerauf DJ, Pentoney AN, Aivati C, Sarakki AV, Doroudgar S, Glembotski CC. Arrieta A, et al. J Biol Chem. 2020 May 29;295(22):7566-7583. doi: 10.1074/jbc.RA120.013345. Epub 2020 Apr 23. J Biol Chem. 2020. PMID: 32327487 Free PMC article. - Post-translational modification: Sweetening protein quality control.
Du Toit A. Du Toit A. Nat Rev Mol Cell Biol. 2014 May;15(5):295. doi: 10.1038/nrm3787. Epub 2014 Apr 3. Nat Rev Mol Cell Biol. 2014. PMID: 24694982 No abstract available. - O-GlcNAc elevation through activation of the hexosamine biosynthetic pathway enhances cancer cell chemoresistance.
Liu Y, Cao Y, Pan X, Shi M, Wu Q, Huang T, Jiang H, Li W, Zhang J. Liu Y, et al. Cell Death Dis. 2018 May 1;9(5):485. doi: 10.1038/s41419-018-0522-0. Cell Death Dis. 2018. PMID: 29706631 Free PMC article.
References
- Champattanachai V, Marchase RB, Chatham JC. Glucosamine protects neonatal cardiomyocytes from ischemia-reperfusion injury via increased protein-associated O-GlcNAc. Am J Physiol Cell Physiol. 2007;292:C178–187. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- DK-099110/DK/NIDDK NIH HHS/United States
- P30 HL101254/HL/NHLBI NIH HHS/United States
- R01 HL102478/HL/NHLBI NIH HHS/United States
- HL-102478-02/HL/NHLBI NIH HHS/United States
- R56 GM038545/GM/NIGMS NIH HHS/United States
- HL-100401/HL/NHLBI NIH HHS/United States
- S10 RR023729/RR/NCRR NIH HHS/United States
- HL-097768/HL/NHLBI NIH HHS/United States
- R01 GM038545/GM/NIGMS NIH HHS/United States
- T32 HL007360/HL/NHLBI NIH HHS/United States
- R01 HL072016/HL/NHLBI NIH HHS/United States
- HL-0980842/HL/NHLBI NIH HHS/United States
- R01 HL120732/HL/NHLBI NIH HHS/United States
- P01 DK088761/DK/NIDDK NIH HHS/United States
- R01 HL097768/HL/NHLBI NIH HHS/United States
- HL-080144/HL/NHLBI NIH HHS/United States
- DK-55758/DK/NIDDK NIH HHS/United States
- DK-088761/DK/NIDDK NIH HHS/United States
- U01 HL100401/HL/NHLBI NIH HHS/United States
- GM-038545/GM/NIGMS NIH HHS/United States
- HL-072016/HL/NHLBI NIH HHS/United States
- R01 HL080144/HL/NHLBI NIH HHS/United States
- R01 HL090842/HL/NHLBI NIH HHS/United States
- R01 DK055758/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous