Single-molecule dynamics of enhanceosome assembly in embryonic stem cells - PubMed (original) (raw)
. 2014 Mar 13;156(6):1274-1285.
doi: 10.1016/j.cell.2014.01.062.
Zhengjian Zhang 2, Li Li 2, Bi-Chang Chen 2, Andrey Revyakin 1, Bassam Hajj 3, Wesley Legant 2, Maxime Dahan 3, Timothée Lionnet 1, Eric Betzig 2, Robert Tjian 4, Zhe Liu 5
Affiliations
- PMID: 24630727
- PMCID: PMC4040518
- DOI: 10.1016/j.cell.2014.01.062
Single-molecule dynamics of enhanceosome assembly in embryonic stem cells
Jiji Chen et al. Cell. 2014.
Abstract
Enhancer-binding pluripotency regulators (Sox2 and Oct4) play a seminal role in embryonic stem (ES) cell-specific gene regulation. Here, we combine in vivo and in vitro single-molecule imaging, transcription factor (TF) mutagenesis, and ChIP-exo mapping to determine how TFs dynamically search for and assemble on their cognate DNA target sites. We find that enhanceosome assembly is hierarchically ordered with kinetically favored Sox2 engaging the target DNA first, followed by assisted binding of Oct4. Sox2/Oct4 follow a trial-and-error sampling mechanism involving 84-97 events of 3D diffusion (3.3-3.7 s) interspersed with brief nonspecific collisions (0.75-0.9 s) before acquiring and dwelling at specific target DNA (12.0-14.6 s). Sox2 employs a 3D diffusion-dominated search mode facilitated by 1D sliding along open DNA to efficiently locate targets. Our findings also reveal fundamental aspects of gene and developmental regulation by fine-tuning TF dynamics and influence of the epigenome on target search parameters.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures
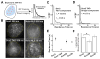
Figure 1. Discriminating Specific and Non-specific TF-DNA Dissociation Kinetics in Live Cells by Single-molecule Imaging
(A) Selective visualization of immobile Sox2 molecules by 2D imaging using long exposure times (low excitation power: 50 W/cm2 and long integration time: 500 ms). Fast particles blend into the background while immobile ones appear as spots. The dissociation rate (koff) is extracted from the measured residence time. (B) Immobile Sox2 molecules imaged with the 2D imaging set-up in ES cells. Top left: Immobile single Sox2 molecules are detected as near diffraction-limited spots in the nucleus. Top right: At 500 ms, halo-tag-NLS (Nuclear Localization Signal) displays negligible stable binding but diffused fluorescence background. Bottom left: A construct lacking the DNA binding domain (Sox2-TAD) displays a great reduction in the number of immobile molecules. Bottom right: At faster frame-rates (10ms), freely diffusing halo-tag-NLS molecules can now be detected as spots. Yellow dotted circle represents the nucleus outline. Scale bar: 2 μm. (C–D) 1-Cumulative Distribution Function (1-CDF) of Sox2 (D) and Sox2-TAD (E) residence time was respectively fitted with a two-component (long-lived and short-lived component) and a single-component exponential decay model. For wtSox2, the fitted lifetimes are _τ_1 = 12.03 ± 1.8 _s (_long-lived component) and _τ_2 = 0.8 ± 0.07 _s (_short-lived component). In the case of Sox2 DNA binding domain deletion (Sox2-TAD), τ = 0.75 ± 0.03 s. (E) Residence lifetime of the long-lived component for Sox2 (12.03 ± 1.8 s), a Sox2 construct with mutations on Sox2-DNA binding surface (Sox2M) (9.11 ± 1.93 s) and Sox2D (8.62 ± 0.98 s) and mean residence lifetime for Sox2-TAD (0.75 ± 0.03 s). (F) Long-lived bound fraction of all bound molecules for Sox2, Sox2M and Sox2D determined by our 2D dwell time analysis. (See Figure S2 and Eq. S1 for details.) *: p<0.05. Error bars represent SD. See also Figures S1 and S2, Movies S1 and S2 and Table S1.
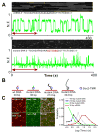
Figure 2. Distinct Sox2 Residence Time Distributions at Specific versus non-specific DNA sequences and Sox2 sliding on DNA measured by in vitro single molecule binding
(A) In vitro single-molecule imaging to determine Sox2 specific and non-specific residence time on DNA. Single-molecule fluorescence traces of halo-TMR Sox2 molecule interacting with surface-tethered wild-type DNA (Upper panel) and mutant DNA (Bottom panel) (N.F. – Normalized Fluorescence) are presented along with the corresponding raw images (The red arrow on the graph indicates the time interval displayed on top of each plot). (B) Schematic of Sox2 interaction with wild type DNA probe (30 bp) and different lengths mutant DNA probes (30 bp, 213 bp and 443 bp) measured by in vitro single molecule imaging. (C) Left panel: Co-localization of wild type DNA probe (Top) or mutant DNA probe (Bottom) (cy5 channel) with TMR-halo-Sox2 molecules (TMR channel). Right panel: histogram of TMR-halo-Sox2 residence time on 30 bp (Green), 213 bp (Blue) and 443 bp mutant DNA (Gray) and 30 bp wild-type (Red). Mean residence time of Sox2 on the 30 bp wild-type DNA is 16.9 s; that on the 30 bp, 213 bp and 443 bp mutant DNA are 0.9 s, 1.6 s and 4.5 s. See also Figure S3, Movie S3 and Table S1.
Figure 3. TF-Chromatin Association Kinetics Determined by Fast 3D Single-molecule Imaging
(A) Fast 3D tracking of TF movement by simultaneous Multi-focus microscopy (axial coverage, 4um; 33Hz). The average association rate (K_on_) is determined through fitting the displacement histogram with a 3D kinetic model (Details see Supplementary information, Eq. S5–13). (B) Volume rendering of 3D Sox2 single-molecule image (purple) superimposed with single-molecule trajectories. Three molecules with distinct behaviors were selectively displayed on the right (from top to bottom: freely diffusing particle; particle undergoing a free/bound transition; immobile molecule). Color bar shows the corresponding frame number. Scale bar: 2 μm. (C) 3D displacement histogram fitted by our 3D kinetic model (Eq. S11) for the indicated factor. Histogram bin was set as 30 nm. The τ_3_D is equal to 1/kon∗ of different TFs was calculated through Eq. S9–12. See also Figure S4, Movie S4 and Table S1.
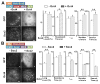
Figure 4. A Trial-and-error TF Target Search Mechanism
(A) Representative ChIP-exo tracks for Sox2 (Antibody), Sox2 (Halo), Sox2D (Halo) and Sox2M (Halo) at two different enhancer regions. Sox2 ChIP-seq data was previously published by Chen et. al (Chen et al., 2008). Auto-scale was applied to each track. (B) Correlation analysis of ChIP-exo data. Upper, enrichment heat-map for all factors ranked by descending order of Sox2 (antibody) enrichment (top 2000 loci). Bottom, heat-map for Sox2 (halo), Sox2D (halo) and Sox2M (halo) showing their own enrichment in the same data set. The read counts within ±100bp regions of exo-peaks were taken into account in this analysis. (C) Calculated duration of the 3D free diffusion state (τ_3_D), the number of trials, the target search time, the ratio that a TF spends in 3D diffusion (3D%) and the number of binding sites determined from genomic analysis (Exo-peak) for Sox2, Sox2D, Sox2M and Oct4. The enrichment cut-off for ChIP-exo peak-pairs is >12 reads for both the left and the right peak (Also see Figure S5C). (D) The model of in vivo target search - a TF goes through on average n episodes of 3D diffusion (τ_3_D = 2.0 ~ 5.6 s, Table S1) interspersed by non-specific binding at random accessible chromatin sites (τns = 0.75 ~ 0.9s, Table S1) before reaching a specific site. τs and τsR denote the search time and the specific residence time, respectively. See also Figures S5 and S6, and Table S1.
Figure 5. Sox2 Assists Oct4 Target Search
(A–B) Sox2 and Oct4 Induction experiment in 3T3 cells. (A), Halo-Sox2 was stably expressed downstream of a CMV promoter. The expression of Oct4 and GFP (IRES) was under the control of an inducible Cumate switch. Thus, GFP is an expression indicator for Oct4. Left: images of Sox2 2D SMT experiments performed with GFP negative/positive cells without or with Oct4 induction. Right: Measured values for the long-lived bound fraction of all bound molecules (from 2D SMT analysis), the specific residence time, the_τ_3_D_, the number of trials and the target search time for Sox2. The results from the converse experiment with Sox2 induction are presented in (B). *: p<0.05, **: p<0.01. Error bars represent SD. See also Movies S5 and S6, and Table S1.
Figure 6. Hierarchically Ordered Enhanceosome Assembly
(A) Structural illustration of mutations (Sox2O and Oct4S) that selectively disrupt Sox2-Oct4 interaction surface. Crystal structures of Sox2 HMG domain (blue) and Oct1 POU domain (red) binding to two different enhancer DNAs (yellow) (FGF4: 1GT0 and UTF1: 1O4X; (Remenyi et al., 2003)) are presented in cartoon model by PyMOL. Mutations on Sox2 (K97E, R100E, M104E and R115E) and reciprocal mutations on Oct4 (I151Y and D159R) are highlighted by purple and green spheres respectively. Notably, these mutations interfere with Sox2-Oct4 interaction in both conformations. (B) Sox2 and Oct4 western blot analysis to examine Sox2 K/D and Oct4 K/D efficiencies in ES cells. Tubulin served as a loading control. (C–D) Changes in the long-lived fraction of all bound molecules, the long-lived residence time, the τ_3_D, the number of trials and the search time of Sox2 and Oct4 after the indicated perturbation experiment. Sox2O and Oct4S as described in (A); Oct4 K/D and Sox2 K/D as described in (B). Error bars represent SD. (E) Sox2 and Oct4 assemble on DNA in an asymmetrically regulated fashion in live cells. We calculated the probability for each reaction route based on Oct4 and Sox2 DNA dissociation and association kinetic information obtained from the 3T3 cell induction experiments (See Eq. S19–26 for calculation details). *: p<0.05, **: p<0.01. See also Table S1.
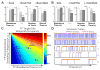
Figure 7. Epigenetic Modulation of TF dynamics and Target Site Occupancy
(A–B) Changes in specific residence time, τ_3_D, number of trials and search time of Sox2 and Oct4 after TSA and 5-AZA treatment in ES cells. ES cells were treated with 50nM TSA for 6 hours or 5μM 5-AZA for 12 hours before the imaging experiment. Error bars represent SD. (C) Simulation heat-map to illustrate the relationship between TF residence time, TF concentration (binding site sampling frequency), and TF target site temporal occupancy. For each pairs of mean TF residence time (x axis) and sampling frequency (y axis), 1000 continuous binding events were simulated and then the temporal occupancy of the target site was calculated based on the total binding On-Off durations. The expected temporal occupancies of Sox2 at specific and non-specific sites derived based on data from Figure S7 were marked with the indicated black lines. The percentage of temporal occupancy is presented in ‘Jet’ color map. See “TF Dynamics Simulation” in the “EXPERIMENTAL PROCEDURES” for details of the parameter set-up. A.U. - Arbitrary Units (D) Individual simulation tracks of distinct TF dynamics regimes (indicated in A) are presented. Specifically, track 1 represents high frequency non-specific chromatin binding. Track 5 represents the TF temporal occupancy of the target site by a TF with a relatively long TF residence time but at low concentrations in the cells. *: p<0.05, **: p<0.01. See also Figure S7 and Table S1.
Similar articles
- Dynamical reorganization of the pluripotency transcription factors Oct4 and Sox2 during early differentiation of embryonic stem cells.
Verneri P, Vazquez Echegaray C, Oses C, Stortz M, Guberman A, Levi V. Verneri P, et al. Sci Rep. 2020 Mar 23;10(1):5195. doi: 10.1038/s41598-020-62235-0. Sci Rep. 2020. PMID: 32251342 Free PMC article. - 3D imaging of Sox2 enhancer clusters in embryonic stem cells.
Liu Z, Legant WR, Chen BC, Li L, Grimm JB, Lavis LD, Betzig E, Tjian R. Liu Z, et al. Elife. 2014 Dec 24;3:e04236. doi: 10.7554/eLife.04236. Elife. 2014. PMID: 25537195 Free PMC article. - A dynamic interplay of enhancer elements regulates Klf4 expression in naïve pluripotency.
Xie L, Torigoe SE, Xiao J, Mai DH, Li L, Davis FP, Dong P, Marie-Nelly H, Grimm J, Lavis L, Darzacq X, Cattoglio C, Liu Z, Tjian R. Xie L, et al. Genes Dev. 2017 Sep 1;31(17):1795-1808. doi: 10.1101/gad.303321.117. Genes Dev. 2017. PMID: 28982762 Free PMC article. - Networks of Transcription Factors for Oct4 Expression in Mice.
Li YQ. Li YQ. DNA Cell Biol. 2017 Sep;36(9):725-736. doi: 10.1089/dna.2017.3780. Epub 2017 Jul 21. DNA Cell Biol. 2017. PMID: 28731785 Review. - The transcriptional foundation of pluripotency.
Chambers I, Tomlinson SR. Chambers I, et al. Development. 2009 Jul;136(14):2311-22. doi: 10.1242/dev.024398. Development. 2009. PMID: 19542351 Free PMC article. Review.
Cited by
- GATA4 Regulates Developing Endocardium Through Interaction With ETS1.
Zhou P, Zhang Y, Sethi I, Ye L, Trembley MA, Cao Y, Akerberg BN, Xiao F, Zhang X, Li K, Jardin BD, Mazumdar N, Ma Q, He A, Zhou B, Pu WT. Zhou P, et al. Circ Res. 2022 Nov 11;131(11):e152-e168. doi: 10.1161/CIRCRESAHA.120.318102. Epub 2022 Oct 20. Circ Res. 2022. PMID: 36263775 Free PMC article. - Real-time single-molecule imaging of transcriptional regulatory networks in living cells.
Hwang DW, Maekiniemi A, Singer RH, Sato H. Hwang DW, et al. Nat Rev Genet. 2024 Apr;25(4):272-285. doi: 10.1038/s41576-023-00684-9. Epub 2024 Jan 9. Nat Rev Genet. 2024. PMID: 38195868 Review. - Mesoscale chromatin confinement facilitates target search of pioneer transcription factors in live cells.
Wang Z, Wang B, Niu D, Yin C, Bi Y, Cattoglio C, Loh KM, Lavis LD, Ge H, Deng W. Wang Z, et al. Nat Struct Mol Biol. 2024 Oct 4. doi: 10.1038/s41594-024-01385-5. Online ahead of print. Nat Struct Mol Biol. 2024. PMID: 39367253 - Sox2 modulation increases naïve pluripotency plasticity.
Tremble KC, Stirparo GG, Bates LE, Maskalenka K, Stuart HT, Jones K, Andersson-Rolf A, Radzisheuskaya A, Koo BK, Bertone P, Silva JCR. Tremble KC, et al. iScience. 2021 Feb 6;24(3):102153. doi: 10.1016/j.isci.2021.102153. eCollection 2021 Mar 19. iScience. 2021. PMID: 33665571 Free PMC article.
References
- Ben-Tabou de-Leon S, Davidson EH. Gene regulation: gene control network in development. Annual review of biophysics and biomolecular structure. 2007;36:191. - PubMed
- Berg OG, Winter RB, von Hippel PH. Diffusion-driven mechanisms of protein translocation on nucleic acids. 1. Models and theory. Biochemistry. 1981;20:6929–6948. - PubMed
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous