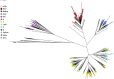PolyTB: a genomic variation map for Mycobacterium tuberculosis - PubMed (original) (raw)
doi: 10.1016/j.tube.2014.02.005. Epub 2014 Feb 15.
Mark Preston 2, José Afonso Guerra-Assunção 3, Grant Hill-Cawthorn 4, David Harris 5, João Perdigão 6, Miguel Viveiros 7, Isabel Portugal 6, Francis Drobniewski 8, Sebastien Gagneux 9, Judith R Glynn 3, Arnab Pain 10, Julian Parkhill 5, Ruth McNerney 2, Nigel Martin 11, Taane G Clark 12
Affiliations
- PMID: 24637013
- PMCID: PMC4066953
- DOI: 10.1016/j.tube.2014.02.005
PolyTB: a genomic variation map for Mycobacterium tuberculosis
Francesc Coll et al. Tuberculosis (Edinb). 2014 May.
Abstract
Tuberculosis (TB) caused by Mycobacterium tuberculosis (Mtb) is the second major cause of death from an infectious disease worldwide. Recent advances in DNA sequencing are leading to the ability to generate whole genome information in clinical isolates of M. tuberculosis complex (MTBC). The identification of informative genetic variants such as phylogenetic markers and those associated with drug resistance or virulence will help barcode Mtb in the context of epidemiological, diagnostic and clinical studies. Mtb genomic datasets are increasingly available as raw sequences, which are potentially difficult and computer intensive to process, and compare across studies. Here we have processed the raw sequence data (>1500 isolates, eight studies) to compile a catalogue of SNPs (n = 74,039, 63% non-synonymous, 51.1% in more than one isolate, i.e. non-private), small indels (n = 4810) and larger structural variants (n = 800). We have developed the PolyTB web-based tool (http://pathogenseq.lshtm.ac.uk/polytb) to visualise the resulting variation and important meta-data (e.g. in silico inferred strain-types, location) within geographical map and phylogenetic views. This resource will allow researchers to identify polymorphisms within candidate genes of interest, as well as examine the genomic diversity and distribution of strains. PolyTB source code is freely available to researchers wishing to develop similar tools for their pathogen of interest.
Keywords: Database; Genomics; Molecular epidemiology; Mycobacterium tuberculosis; Software; Whole-genome sequencing.
Copyright © 2014 The Authors. Published by Elsevier Ltd.. All rights reserved.
Figures
Figure 1
RAxML maximum likelihood phylogenetic tree constructed for all 1470 isolates (spoligotype colour-coded). Radial phylogram representation of the best-scoring maximum likelihood phylogenetic tree constructed using RAxML software. Samples are colour-coded by spoligotype strain showing a clear correlation of SNP and spoligotype clustering.
Figure 2
Polymorphism frequency and density plots. (a) SNP frequency bar plot; (b) SNP density plots for all, coding, non-coding and Tuberculist-functional annotated families; (c) Small indel frequency plot; (d) Indel density plots for all, coding, non-coding and functional-grouped coding indels.
Figure 3
Polymorphisms at the rpoB-rpoC region associated with rifampicin resistance (Browser View). Genetic variants are shown at the _rpoB a_nd rpoC genes, loci known to be associated with rifampicin resistance. Synonymous SNPs (sSNPs) are coloured in black, non-synonymous SNPs (nsSNPs) in red and small insertions and deletions in blue and green, respectively. Cursor movement over variants displays an information box with further annotation including nucleotide, codon and amino acid changes for SNPs; and length and sequence for indels. Locations and Spoligotypes tracks are placed as colour-coded vertical bars at the left hand side of the genomic plot and provide information for samples. Sixty isolates are shown, 30 from Malawi (colour-coded in red in the Location bar) and 30 from Uganda (shown in green). Patterns of SNP difference can be observed when comparing isolates from different populations: Kampala isolates harbour many more nsSNPs at rpoB gene than Malawian isolates. The observed nsSNPs are likely to be the underlying cause of rifampicin resistance (Clark et al., 2013). In fact, _rpoB-_516 (A → T SNP at 761,110 bp), rpoB-526 (G → T 761,139 bp and A → G 761,140 bp) and _rpoB-_531 (C → G 761,155 bp) mutations are observed in Ugandan isolates, and correspond to nsSNPs already reported as rifampicin resistance markers .
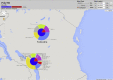
Figure 4
SNP associated with lineage 1 (EAI) in Tanzanian and Malawian populations (Map view). Allele frequencies are shown for the chosen polymorphic position as pie charts, either alone or combined with in silico inferred spoligotypes (Coll et al., 2012) to allow the visual detection of relationships between certain alleles and strain types. Reference allele frequency portions on pie charts are coloured in blue while alternative allele (i.e. non-reference) frequencies are shown in red. Outer chart portions representing relative strain type frequencies are colour-coded by main spoligotype families (AFRI, BOV, Beijing, CAS, EAI, LAM, Manu, S, T and X). In this particular case, the SNP at 4,411,016 bp position is found to be associated with lineage 1 (EAI) strains in Tanzania and Karonga (Malawi) populations, visualised as the red portion of the inner pie chart linking with the purple portions of the outer pie in both settings.
Figure 5
SNP-based neighbour-joining phylogenetic tree of 140 isolates belonging to four different locations (Phylogenetic view) A neighbour phylogenetic tree based on pre-calculated SNP distances is built in real time for the set of 140 isolates from Shanghai (China), Hamburg (Germany), Karonga (Malawi) and Kampala (Uganda). Spoligotype lineages and locations are colour-coded as bar charts around the tree (outer bar representing locations and the inner one spoligotypes) to enable the visual identification of correlations between spoligotype/location and phylogenetic clustering. A table summarising all colour codes will be shown at the left hand side of the page.
Similar articles
- PhyTB: Phylogenetic tree visualisation and sample positioning for M. tuberculosis.
Benavente ED, Coll F, Furnham N, McNerney R, Glynn JR, Campino S, Pain A, Mohareb FR, Clark TG. Benavente ED, et al. BMC Bioinformatics. 2015 May 13;16(1):155. doi: 10.1186/s12859-015-0603-3. BMC Bioinformatics. 2015. PMID: 25968323 Free PMC article. - [Future prospects of molecular epidemiology in tuberculosis].
Matsumoto T, Iwamoto T. Matsumoto T, et al. Kekkaku. 2009 Dec;84(12):783-4. Kekkaku. 2009. PMID: 20077862 Japanese. - Whole Genome Sequencing of Mycobacterium tuberculosis Isolates From Extrapulmonary Sites.
Sharma K, Verma R, Advani J, Chatterjee O, Solanki HS, Sharma A, Varma S, Modi M, Ray P, Mukherjee KK, Sharma M, Dhillion MS, Suar M, Chatterjee A, Pandey A, Prasad TSK, Gowda H. Sharma K, et al. OMICS. 2017 Jul;21(7):413-425. doi: 10.1089/omi.2017.0070. OMICS. 2017. PMID: 28692415 Free PMC article. - Bioinformatics tools and databases for whole genome sequence analysis of Mycobacterium tuberculosis.
Faksri K, Tan JH, Chaiprasert A, Teo YY, Ong RT. Faksri K, et al. Infect Genet Evol. 2016 Nov;45:359-368. doi: 10.1016/j.meegid.2016.09.013. Epub 2016 Sep 13. Infect Genet Evol. 2016. PMID: 27637931 Review. - Large genomics datasets shed light on the evolution of the Mycobacterium tuberculosis complex.
Chiner-Oms Á, Comas I. Chiner-Oms Á, et al. Infect Genet Evol. 2019 Aug;72:10-15. doi: 10.1016/j.meegid.2019.02.028. Epub 2019 Feb 26. Infect Genet Evol. 2019. PMID: 30822550 Review.
Cited by
- Insertion and deletion evolution reflects antibiotics selection pressure in a Mycobacterium tuberculosis outbreak.
Godfroid M, Dagan T, Merker M, Kohl TA, Diel R, Maurer FP, Niemann S, Kupczok A. Godfroid M, et al. PLoS Pathog. 2020 Sep 30;16(9):e1008357. doi: 10.1371/journal.ppat.1008357. eCollection 2020 Sep. PLoS Pathog. 2020. PMID: 32997707 Free PMC article. - SNPs in genes related to the repair of damage to DNA in clinical isolates of M. tuberculosis: A transversal and longitudinal approach.
Pérez-Martínez DE, Zenteno-Cuevas R. Pérez-Martínez DE, et al. PLoS One. 2024 Jun 25;19(6):e0295464. doi: 10.1371/journal.pone.0295464. eCollection 2024. PLoS One. 2024. PMID: 38917091 Free PMC article. - CRISPRbuilder-TB: "CRISPR-builder for tuberculosis". Exhaustive reconstruction of the CRISPR locus in mycobacterium tuberculosis complex using SRA.
Guyeux C, Sola C, Noûs C, Refrégier G. Guyeux C, et al. PLoS Comput Biol. 2021 Mar 5;17(3):e1008500. doi: 10.1371/journal.pcbi.1008500. eCollection 2021 Mar. PLoS Comput Biol. 2021. PMID: 33667225 Free PMC article. - Recombination in pe/ppe genes contributes to genetic variation in Mycobacterium tuberculosis lineages.
Phelan JE, Coll F, Bergval I, Anthony RM, Warren R, Sampson SL, Gey van Pittius NC, Glynn JR, Crampin AC, Alves A, Bessa TB, Campino S, Dheda K, Grandjean L, Hasan R, Hasan Z, Miranda A, Moore D, Panaiotov S, Perdigao J, Portugal I, Sheen P, de Oliveira Sousa E, Streicher EM, van Helden PD, Viveiros M, Hibberd ML, Pain A, McNerney R, Clark TG. Phelan JE, et al. BMC Genomics. 2016 Feb 29;17:151. doi: 10.1186/s12864-016-2467-y. BMC Genomics. 2016. PMID: 26923687 Free PMC article. - Unexpected diversity of CRISPR unveils some evolutionary patterns of repeated sequences in Mycobacterium tuberculosis.
Refrégier G, Sola C, Guyeux C. Refrégier G, et al. BMC Genomics. 2020 Nov 30;21(1):841. doi: 10.1186/s12864-020-07178-6. BMC Genomics. 2020. PMID: 33256602 Free PMC article.
References
- Abubakar I., Zignol M., Falzon D., Raviglione M., Ditiu L., Masham S., Adetifa I., Ford N., Cox H., Lawn S.D., Marais B.J., McHugh T.D., Mwaba P., Bates M., Lipman M., Zijenah L., Logan S., McNerney R., Zumla A., Sarda K., Nahid P., Hoelscher M., Pletschette M., Memish Z.a., Kim P., Hafner R., Cole S., Migliori G.B., Maeurer M., Schito M., Zumla A. Drug-resistant tuberculosis: time for visionary political leadership. Lancet Infect Dis. 2013;13:529–539. - PubMed
- Garcia-Betancur J.C., Menendez M.C., Del Portillo P., Garcia M.J. Alignment of multiple complete genomes suggests that gene rearrangements may contribute towards the speciation of Mycobacteria. Infect Genet Evol. 2011;12:819–826. - PubMed
- Blouin Y., Hauck Y., Soler C., Fabre M., Vong R., Dehan C., Cazajous G., Massoure P.-L., Kraemer P., Jenkins A., Garnotel E., Pourcel C., Vergnaud G. Significance of the identification in the horn of Africa of an exceptionally deep branching Mycobacterium tuberculosis clade. PloS One. 2012;7:e52841. - PMC - PubMed
- Comas I., Coscolla M., Luo T., Borrell S., Holt K.E., Kato-Maeda M., Parkhill J., Malla B., Berg S., Thwaites G., Yeboah-Manu D., Bothamley G., Mei J., Wei L., Bentley S., Harris S.R., Niemann S., Diel R., Aseffa A., Gao Q., Young D., Gagneux S. Out-of-Africa migration and neolithic coexpansion of Mycobacterium tuberculosis with modern humans. Nat Genet. 2013;45:1176–1182. - PMC - PubMed
- Supply P., Marceau M., Mangenot S., Roche D., Rouanet C., Khanna V., Majlessi L., Criscuolo A., Tap J., Pawlik A., Fiette L., Orgeur M., Fabre M., Parmentier C., Frigui W., Simeone R., Boritsch E.C., Debrie A.-S., Willery E., Walker D., Quail M.a., Ma L., Bouchier C., Salvignol G., Sayes F., Cascioferro A., Seemann T., Barbe V., Locht C., Gutierrez M.-C., Leclerc C., Bentley S.D., Stinear T.P., Brisse S., Médigue C., Parkhill J., Cruveiller S., Brosch R. Genomic analysis of smooth tubercle bacilli provides insights into ancestry and pathoadaptation of Mycobacterium tuberculosis. Nat Genet. 2013;45:172–179. - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical