PMA induces SnoN proteolysis and CD61 expression through an autocrine mechanism - PubMed (original) (raw)
PMA induces SnoN proteolysis and CD61 expression through an autocrine mechanism
Chonghua Li et al. Cell Signal. 2014 Jul.
Abstract
Phorbol-12-myristate-13-acetate, also called PMA, is a small molecule that activates protein kinase C and functions to differentiate hematologic lineage cells. However, the mechanism of PMA-induced cellular differentiation is not fully understood. We found that PMA triggers global enhancement of protein ubiquitination in K562, a myelogenous leukemia cell line and one of the enhanced-ubiquitination targets is SnoN, an inhibitor of the Smad signaling pathway. Our data indicated that PMA stimulated the production of Activin A, a cytokine of the TGF-β family. Activin A then activated the phosphorylation of both Smad2 and Smad3. In consequence, SnoN is ubiquitinated by the APC(Cdh1) ubiquitin ligase with the help of phosphorylated Smad2. Furthermore, we found that SnoN proteolysis is important for the expression of CD61, a marker of megakaryocyte. These results indicate that protein ubiquitination promotes megakaryopoiesis via degrading SnoN, an inhibitor of CD61 expression, strengths the roles of ubiquitination in cellular differentiation.
Keywords: Activin A; Cdh1; Differentiation; Smad2; SnoN; Ubiquitin.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures
Figure 1
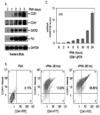
PMA induces megakaryopoiesis of K562 cells. Cells were treated with PMA and collected at different time points. (A). Analyze expressions of CD41, CD61, GATA2 and Fli-1 using western blots. (B). Analyze CD41 and CD61 expression using Flow cytometry. (C). Analyze CD61 expression using qPCR.
Figure 2
The ubiquitination machinery is altered upon PMA treatment. K562-His-Bio-Ub or K562 cells were treated with PMA and collected at different time points. (A). Overall ubiquitination was enhanced by PMA. K562-His-Bio-Ub cells were collected after 4 and 8 hours of PMA treatment. Ubiquitinated proteins were purified using Streptavidin beads under denatured conditions and subjected to SDS-PAGE electrophoresis. α-Ubiquitin (FK2) antibody was employed to detect ubiquitinated proteins. (B). Different Ubiquitin-related genes were regulated by PMA. K562 cells were collected at different days post PMA treatment and subjected to SDS-PAGE electrophoresis. Antibodies against various ubiquitin ligases were used for western blots.
Figure 3
SnoN is degraded via the ubiquitin and proteasome pathway upon PMA treatment. Western blots were employed to detect protein expression. (A). SnoN was down-regulated by PMA. (B). MG132 inhibited SnoN degradation. K562 cells were treated with PMA for 24 hours. MG132 at 20 µM was added to block protein degradation. (C). Bortezomib blocked SnoN turnover. K562 cells were treated with PMA for 24 hours. Bortezomib at 20 µM was added to block protein degradation. (D). SnoN was ubiquitinated. K562-Bio-Ub cells were collected after 8 hours of PMA treatment. Ubiquitinated proteins were purified using Streptavidin beads and subjected to SDS-PAGE electrophoresis. α-SnoN antibody was employed to detect ubiquitinated SnoN. (E). Exogenous Bio-SnoN responded to PMA treatment. K562 and K562-Bio-SnoN cells were treated with PMA for 24 hours. MG132 at 40 µM was added to block SnoN turnover 2 hours before sample collection. Bio-SnoN was detected using Streptavidin-HRP. (F). K48 polyubiquitin chain was identified on SnoN. Collected cells from Figure 3E were lysed in a denaturing buffer and Bio-SnoN proteins were purified using Streptavidin beads. After resolving on a SDS-PAGE gel, ubiquitinated Bio-SnoN proteins were detected using ant-K48 polyubiquitin chain-specific antibody (upper panel). An – SnoN antibody was employed to detect Bio-SnoN (low panel).
Figure 4
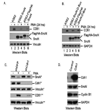
Cdh1 is important for PMA-induced proteolysis of SnoN whose over-expression could attenuate CD61 induction. Both untagged and Flag/HA-tagged SnoN were expressed in K562 cells. PMA was used to trigger SnoN turnover and CD61 expression. Cells were collected at 7 or 24 hours post PMA treatment. ShRNA targeting Cdh1 was expressed using lentivirus system. Western blots were employed to detect protein expressions. (A). Exogenous SnoN inhibits CD61 induction. (B). The D-box mutant of SnoN was more stable and could block CD61 induction. (C). Silencing Cdh1 inhibited SnoN turnover and CD61 induction. Endogenous Cdh1 was silenced using lentivirus expressing shRNA in K562 cells. Cells were collected at 7 hours (*) post PMA treatment to detect SnoN, Cdh1 and Vinculin. Cells were also collected at 24 hours (**) post PMA treatment for better results of CD61 induction using western blots. (D). Over-expression of Cdh1 could reduce Cyclin B1, but not SnoN expression. Exogenous Cdh1 was over-expressed in K562 cells using lentivirus approach. Infected cells were subjected to western blots.
Figure 5
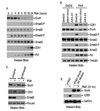
Activated Smad2 signaling is required for SnoN turnover and CD61 induction. (A). Both Smad2 and Smad3 were phosphorylated upon PMA treatment. K562 cells were treated with PMA for different times. Western blots were employed to detect expression of Smad2, Smad3, Phospho-Smad2 (Ser465/467) and Phospho-Smad3 (Ser423/425). (B). Small molecule inhibitors of TGFβ type I receptors blocked SnoN proteolysis and CD61 expression. SB413542 (10µM) or A83-01 (1µM) was added at the same time as PMA was done in K562 cells. Cells were collected 24 hours later and subjected to western blots. (C). Knocking down Smad2 inhibited SnoN turnover and CD61 expression. Endogenous Smad2 was silenced using lentivirus expressing shRNA in K562 cells. Cells were collected at 8 hours post PMA treatment and subjected to SDS-PAGE electrophoresis and western blots. (D). The SnoN A3 mutant inhibited CD61 induction. Threonine-89, Leucine-90 and Glutamine-92 of SnoN were mutated to Alanines as a SnoN A3 mutant (T89A/L90A/Q92A). The untagged A3 mutant was expressed in K562 cells using lentivirus infection. Expression of SnoN and CD61 were detected using western blot.
Figure 6
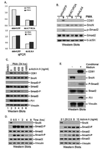
The Activin A-ACVR2A-Alk4 signaling axis is activated upon PMA treatment. K562 cells were collected at different time points after PMA treatment for cDNA preparation using reverse transcription. (A). Expression of Alk4 was examined using qPCR. (B). Expression of ACVR2A was examined using qPCR. (C). Expression of Activin A was examined using qPCR. (D). Activin A proteins were secreted into the culture media after PMA treatment. Culture media were collected at different time points after PMA treatment. Activin A proteins were detected using ELISA.
Figure 7
Activin A and its receptors play important roles in PMA-induced megakaryopoiesis. (A). ACVR2A or Alk4 was silenced in K562 cells using shRNA. qPCR was used to show knockdown efficiency. (B). Silencing either ACVR2A or Alk4 could attenuate PMA-stimulated Smad2 phosphorylation, SnoN degradation and CD61 induction. Control and knockdown cells were treated with PMA for 8 hours and cells were collected for western blots. (C). Activin A antibody could attenuate PMA-induced megakaryopoiesis. Different amounts of Activin A antibodies were used to treat K562 cells at the same time as PMA. 24 hours later, cells were collected for western blots. (D). Recombinant Activin A proteins triggered Smad signaling pathway and SnoN turnover. Left Panel: Time course of Activin A treatment. Recombinant Activin A proteins (10 ng/ml) were employed to treat K562 cells. Cells were collected at different time points for western blots. Right Panel: Dosage effect of Activin A. Different amounts of recombinant Activin A proteins were employed to treat K562 cells for 8 hours. Cells were collected for western blots. (E). Conditional media could trigger the similar effect to PMA treatment. K562 cells were treated with either DMSO or PMA for 3 hours. PMA was then washed away with PBS and cultured for another 5 hours without PMA or DMSO to produce conditional media. Conditioned media were collected and used to treat K562 cells for 24 hours. Cells were collected for western blots.
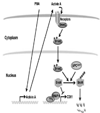
Figure 8
Model of PMA-induced SnoN turnover and CD61 expression. PMA stimulates expression of Activin A which activates Smad2 signaling pathway via activating its receptors, such as Alk4. Phosphorylated Smad2 functions as a cofactor for the APCCdh1 ubiquitin ligase to ubiquitinate SnoN. SnoN functions as an inhibitor of CD61 expression and its proteolysis via the ubiquitin and proteasome pathway is important for PMA-induced CD61 induction.
Similar articles
- Arkadia activates Smad3/Smad4-dependent transcription by triggering signal-induced SnoN degradation.
Levy L, Howell M, Das D, Harkin S, Episkopou V, Hill CS. Levy L, et al. Mol Cell Biol. 2007 Sep;27(17):6068-83. doi: 10.1128/MCB.00664-07. Epub 2007 Jun 25. Mol Cell Biol. 2007. PMID: 17591695 Free PMC article. - Smad3 recruits the anaphase-promoting complex for ubiquitination and degradation of SnoN.
Stroschein SL, Bonni S, Wrana JL, Luo K. Stroschein SL, et al. Genes Dev. 2001 Nov 1;15(21):2822-36. doi: 10.1101/gad.912901. Genes Dev. 2001. PMID: 11691834 Free PMC article. - Downregulation of SnoN expression in obstructive nephropathy is mediated by an enhanced ubiquitin-dependent degradation.
Tan R, Zhang J, Tan X, Zhang X, Yang J, Liu Y. Tan R, et al. J Am Soc Nephrol. 2006 Oct;17(10):2781-91. doi: 10.1681/ASN.2005101055. Epub 2006 Sep 7. J Am Soc Nephrol. 2006. PMID: 16959829 - SnoN in TGF-beta signaling and cancer biology.
Pot I, Bonni S. Pot I, et al. Curr Mol Med. 2008 Jun;8(4):319-28. doi: 10.2174/156652408784533797. Curr Mol Med. 2008. PMID: 18537639 Review.
Cited by
- Transcriptional cofactors Ski and SnoN are major regulators of the TGF-β/Smad signaling pathway in health and disease.
Tecalco-Cruz AC, Ríos-López DG, Vázquez-Victorio G, Rosales-Alvarez RE, Macías-Silva M. Tecalco-Cruz AC, et al. Signal Transduct Target Ther. 2018 Jun 8;3:15. doi: 10.1038/s41392-018-0015-8. eCollection 2018. Signal Transduct Target Ther. 2018. PMID: 29892481 Free PMC article. - The Role of the APC/C and Its Coactivators Cdh1 and Cdc20 in Cancer Development and Therapy.
Greil C, Engelhardt M, Wäsch R. Greil C, et al. Front Genet. 2022 Jun 27;13:941565. doi: 10.3389/fgene.2022.941565. eCollection 2022. Front Genet. 2022. PMID: 35832196 Free PMC article. Review. - Suppression of APC/CCdh1 has subtype specific biological effects in acute myeloid leukemia.
Ewerth D, Schmidts A, Hein M, Schnerch D, Kvainickas A, Greil C, Duyster J, Engelhardt M, Wäsch R. Ewerth D, et al. Oncotarget. 2016 Jul 26;7(30):48220-48230. doi: 10.18632/oncotarget.10196. Oncotarget. 2016. PMID: 27374082 Free PMC article. - The Involvement of Canonical NFκB Pathway in Megakaryocyte Differentiation Induction by Nanocurcumin.
Mortazavi Farsani SS, Sadeghizadeh D, Babashah S, Rad F, Sadeghizadeh M. Mortazavi Farsani SS, et al. Int J Hematol Oncol Stem Cell Res. 2023 Jan 1;17(1):18-27. doi: 10.18502/ijhoscr.v17i1.11709. Int J Hematol Oncol Stem Cell Res. 2023. PMID: 37638286 Free PMC article.
References
- Pickart CM. Back to the future with ubiquitin. Cell. 2004;116:181–190. - PubMed
- Harper JW, Elledge SJ. Mol. Cell. 2007;28:739–745. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous