Function, therapeutic potential and cell biology of BACE proteases: current status and future prospects - PubMed (original) (raw)
Review
Function, therapeutic potential and cell biology of BACE proteases: current status and future prospects
Robert Vassar et al. J Neurochem. 2014 Jul.
Abstract
The β-site APP cleaving enzymes 1 and 2 (BACE1 and BACE2) were initially identified as transmembrane aspartyl proteases cleaving the amyloid precursor protein (APP). BACE1 is a major drug target for Alzheimer's disease because BACE1-mediated cleavage of APP is the first step in the generation of the pathogenic amyloid-β peptides. BACE1, which is highly expressed in the nervous system, is also required for myelination by cleaving neuregulin 1. Several recent proteomic and in vivo studies using BACE1- and BACE2-deficient mice demonstrate a much wider range of physiological substrates and functions for both proteases within and outside of the nervous system. For BACE1 this includes axon guidance, neurogenesis, muscle spindle formation, and neuronal network functions, whereas BACE2 was shown to be involved in pigmentation and pancreatic β-cell function. This review highlights the recent progress in understanding cell biology, substrates, and functions of BACE proteases and discusses the therapeutic options and potential mechanism-based liabilities, in particular for BACE inhibitors in Alzheimer's disease. The protease BACE1 is a major drug target in Alzheimer disease. Together with its homolog BACE2, both proteases have an increasing number of functions within and outside of the nervous system. This review highlights recent progress in understanding cell biology, substrates, and functions of BACE proteases and discusses the therapeutic options and potential mechanism-based liabilities, in particular for BACE inhibitors in Alzheimer disease.
Keywords: Alzheimer's disease; BACE1; BACE2; protease; regulated intramembrane proteolysis; secretase.
© 2014 International Society for Neurochemistry.
Figures
Fig. 1
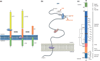
APP processing, FAD mutations, and β-site APP cleaving enzyme (BACE)1. (a) APP is a type-I membrane protein that is sequentially cleaved by two aspartic proteases to generate Aβ. First, the β-secretase enzyme (β) cuts APP (1) to create the N-terminus of Aβ. Two APP fragments are produced: membrane-bound C99 and secreted sAPPβ ectodomain (yellow). Second, C99 is cleaved by the γ-secretase enzyme (γ) to generate the C-terminus of Aβ. Aβ (orange) is then released into the lumen of the endosome and secreted into the extracellular medium. An intracellular domain, C59 (green), is also produced. (b) The membrane-bound APP polypeptide is represented by the gray string. APP residues that affect β-secretase processing of APP in humans are represented by gray circles, within which the wild-type residue is identified by the single-letter amino acid code. The K670N/M671L (Swedish) and A673V mutations cause FAD by increasing the rate of β-secretase cleavage and Aβ production, whereas the A673T mutation protects against Alzheimer’s disease (AD) by doing the opposite. All three mutations occur at or within one amino acid of the β-secretase cleavage site. Red, blue, and lavender notched ellipses represent α, β, and γ-secretases, respectively, cutting at their respective cleavage sites in APP. (c) BACE1 is a 501 amino acid type-I transmembrane aspartic protease. The various subdomains of BACE1 are indicated to the right of the structure. Numbers and letters refer to amino acid positions and single-letter code, respectively. The two signature aspartic protease active site motifs at positions 93 and 289 are shaded black. S–S denote positions of disulfide bridges within the catalytic domain; N represents positions of N-linked glycosylation sites; R indicates positions of acetylated arginine residues; C marks positions of _S_-palmitoylated cysteine residues; P indicates phosphorylation of serine 498; Ub denotes ubiquitination of lysine 501.
Fig. 2
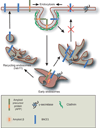
Beta-site amyloid cleaving enzyme (BACE)1 trafficking requires recycling endosomes. BACE1 traffics from plasma membrane to early endosomes where it cleaves most of the cellular APP. From the early endosomes, BACE1 is routed to Rab11-GTPase positive recycling endosomes, through which it is sorted to plasma membrane to reinitiate another round of entry into early endosomes to cleave APP. In the absence of functional Rab11 GTPase, much of BACE1 accumulates in recycling compartments and fails to be recycled to early endosomes, as a result of which β-cleavage of APP and Aβ production are significantly decreased.
Fig. 3
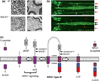
Beta-site amyloid cleaving enzyme (BACE)1 controls peripheral nerve myelination and muscle spindle formation via proteolytic processing of neuregulin 1. (a) BACE1 deficiency results in hypomyelination of peripheral axons (left panel) and abnormalities in axonal-bundling (right panel) (Willem et al. 2006). (b) Delay of myelination in BACE1−/− zebrafish (van Bebber et al. 2013). At 3 days post-fertilization myelination of the posterior lateral line nerves (PNS) is severely reduced in BACE1−/−; claudin k: GFP (red arrows) whereas CNS derived oligodendrocytes ensheathing the Mauthner axons are normally myelinated (black arrows). (c) Proteolytic processing of neuregulin 1 (NRG1) type III by sheddases and intramembrane proteolysis.
Fig. 4
Beta-site amyloid cleaving enzyme (BACE)2 regulates pigmentation in mice and melanophore migration in zebrafish. (a) Deletion of Bace2 in mice leads to dilution of coat color (Rochin et al. 2013). Note the black coat normally seen in the C57Bl/6 strain as observed in control and Bace1 knockout mice. (b) Deletion of Bace2 in zebrafish alters the shape and migration of melanophores (van Bebber et al. 2013). Whole fish (left panels); boxed areas are enlarged to show details near end of yolk sac extension (middle panels) and fin (right panels).
Fig. 5
(a) Example of an optimized peptidomimetic β-site amyloid cleaving enzyme (BACE) inhibitor with high molecular weight and marginal in vivo Aβ lowering activity at very high doses of 250 mg/kg (Hussain et al. 2007). (b). Optimized cyclic isothiourea BACE inhibitor derived from a fragment-based screening approach (May et al. 2011). LY2811376 displays modest BACE1 potency but with robust in vivo Aβ lowering activity in animals and normal healthy volunteers in Phase 1 studies. LY2811376 development was terminated during Phase 1 owing to off target toxicities in eye and brain. (c) Merck iminoheterocyclic BACE inhibitor series evolved from a low affinity isothiourea hit (Merck D) identified in a protein NMR-based screen. Replacement off the isothiourea warhead with the more drug-like iminohydantoin core (Merck E) led to further optimization (Merck G) and ring expansion of the warhead to produce molecules like Merck J with several orders of magnitude improvements in intrinsic affinity with relatively small increase in molecular weight. MK-8931 builds upon the favorable properties described here for the iminoheterocyclic BACE inhibitor series with significant improvements in intrinsic affinity, selectivity, in vivo potency, and other key properties.
Similar articles
- Beta-secretase (BACE) as a drug target for Alzheimer's disease.
Vassar R. Vassar R. Adv Drug Deliv Rev. 2002 Dec 7;54(12):1589-602. doi: 10.1016/s0169-409x(02)00157-6. Adv Drug Deliv Rev. 2002. PMID: 12453676 Review. - The beta-secretase, BACE: a prime drug target for Alzheimer's disease.
Vassar R. Vassar R. J Mol Neurosci. 2001 Oct;17(2):157-70. doi: 10.1385/JMN:17:2:157. J Mol Neurosci. 2001. PMID: 11816789 Review. - BACE1: the beta-secretase enzyme in Alzheimer's disease.
Vassar R. Vassar R. J Mol Neurosci. 2004;23(1-2):105-14. doi: 10.1385/JMN:23:1-2:105. J Mol Neurosci. 2004. PMID: 15126696 Review. - BACE1 Physiological Functions May Limit Its Use as Therapeutic Target for Alzheimer's Disease.
Barão S, Moechars D, Lichtenthaler SF, De Strooper B. Barão S, et al. Trends Neurosci. 2016 Mar;39(3):158-169. doi: 10.1016/j.tins.2016.01.003. Epub 2016 Jan 30. Trends Neurosci. 2016. PMID: 26833257 Review. - Distinct transcriptional regulation and function of the human BACE2 and BACE1 genes.
Sun X, Wang Y, Qing H, Christensen MA, Liu Y, Zhou W, Tong Y, Xiao C, Huang Y, Zhang S, Liu X, Song W. Sun X, et al. FASEB J. 2005 May;19(7):739-49. doi: 10.1096/fj.04-3426com. FASEB J. 2005. PMID: 15857888
Cited by
- Current and future implications of basic and translational research on amyloid-β peptide production and removal pathways.
Bohm C, Chen F, Sevalle J, Qamar S, Dodd R, Li Y, Schmitt-Ulms G, Fraser PE, St George-Hyslop PH. Bohm C, et al. Mol Cell Neurosci. 2015 May;66(Pt A):3-11. doi: 10.1016/j.mcn.2015.02.016. Epub 2015 Mar 4. Mol Cell Neurosci. 2015. PMID: 25748120 Free PMC article. Review. - Functions of the Alzheimer's Disease Protease BACE1 at the Synapse in the Central Nervous System.
Munro KM, Nash A, Pigoni M, Lichtenthaler SF, Gunnersen JM. Munro KM, et al. J Mol Neurosci. 2016 Nov;60(3):305-315. doi: 10.1007/s12031-016-0800-1. Epub 2016 Jul 25. J Mol Neurosci. 2016. PMID: 27456313 Free PMC article. Review. - Significance of transcytosis in Alzheimer's disease: BACE1 takes the scenic route to axons.
Buggia-Prévot V, Thinakaran G. Buggia-Prévot V, et al. Bioessays. 2015 Aug;37(8):888-98. doi: 10.1002/bies.201500019. Epub 2015 Jun 30. Bioessays. 2015. PMID: 26126792 Free PMC article. - The potent BACE1 inhibitor LY2886721 elicits robust central Aβ pharmacodynamic responses in mice, dogs, and humans.
May PC, Willis BA, Lowe SL, Dean RA, Monk SA, Cocke PJ, Audia JE, Boggs LN, Borders AR, Brier RA, Calligaro DO, Day TA, Ereshefsky L, Erickson JA, Gevorkyan H, Gonzales CR, James DE, Jhee SS, Komjathy SF, Li L, Lindstrom TD, Mathes BM, Martényi F, Sheehan SM, Stout SL, Timm DE, Vaught GM, Watson BM, Winneroski LL, Yang Z, Mergott DJ. May PC, et al. J Neurosci. 2015 Jan 21;35(3):1199-210. doi: 10.1523/JNEUROSCI.4129-14.2015. J Neurosci. 2015. PMID: 25609634 Free PMC article. Clinical Trial. - Potential Benefits of Nobiletin, A Citrus Flavonoid, against Alzheimer's Disease and Parkinson's Disease.
Nakajima A, Ohizumi Y. Nakajima A, et al. Int J Mol Sci. 2019 Jul 10;20(14):3380. doi: 10.3390/ijms20143380. Int J Mol Sci. 2019. PMID: 31295812 Free PMC article. Review.
References
- Akpinar P, Kuwajima S, Krutzfeldt J, Stoffel M. Tmem27: a cleaved and shed plasma membrane protein that stimulates pancreatic beta cell proliferation. Cell Metab. 2005;2:385–397. - PubMed
- Albert JS. Progress in the development of beta-secretase inhibitors for Alzheimer’s disease. Prog. Med. Chem. 2009;48:133–161. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P50 AG005146/AG/NIA NIH HHS/United States
- RF1 AG022560/AG/NIA NIH HHS/United States
- R01 AG030142/AG/NIA NIH HHS/United States
- R01AG022560/AG/NIA NIH HHS/United States
- R01 AG022560/AG/NIA NIH HHS/United States
- R01AG030142/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources