Human oral viruses are personal, persistent and gender-consistent - PubMed (original) (raw)
Human oral viruses are personal, persistent and gender-consistent
Shira R Abeles et al. ISME J. 2014 Sep.
Abstract
Viruses are the most abundant members of the human oral microbiome, yet relatively little is known about their biodiversity in humans. To improve our understanding of the DNA viruses that inhabit the human oral cavity, we examined saliva from a cohort of eight unrelated subjects over a 60-day period. Each subject was examined at 11 time points to characterize longitudinal differences in human oral viruses. Our primary goals were to determine whether oral viruses were specific to individuals and whether viral genotypes persisted over time. We found a subset of homologous viral genotypes across all subjects and time points studied, suggesting that certain genotypes may be ubiquitous among healthy human subjects. We also found significant associations between viral genotypes and individual subjects, indicating that viruses are a highly personalized feature of the healthy human oral microbiome. Many of these oral viruses were not transient members of the oral ecosystem, as demonstrated by the persistence of certain viruses throughout the entire 60-day study period. As has previously been demonstrated for bacteria and fungi, membership in the oral viral community was significantly associated with the sex of each subject. Similar characteristics of personalized, sex-specific microflora could not be identified for oral bacterial communities based on 16S rRNA. Our findings that many viruses are stable and individual-specific members of the oral ecosystem suggest that viruses have an important role in the human oral ecosystem.
Figures
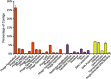
Figure 1
Bar graph of the percentage of contigs (±s.e.) with viral homologs in the NR database from all subjects.
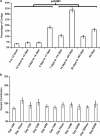
Figure 2
Bar graph (±s.e.) demonstrating the relative time intervals between the earliest and latest time points that formed each contig (a) and the relative contribution of each time point to contigs formed from all 11 time points (b). Significance testing was determined by two-tailed _t_-test.
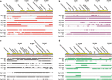
Figure 3
Assemblies of contigs from all time points in each subject. (a) Viral contig from subject no. 1, (b) subject no. 2, (c) subject no. 3, (d) subject no. 4, (e) subject no. 5, (f) subject no. 6, (g) subject no. 7 and (h) subject no. 8. The portions of the contigs identified at each time point are represented by the colored boxes for each subject. The relative coverage of each contig is represented, along with annotated open reading frames (ORFs) above each diagram. The length of each contig is denoted at the top of each panel.
Figure 3
Assemblies of contigs from all time points in each subject. (a) Viral contig from subject no. 1, (b) subject no. 2, (c) subject no. 3, (d) subject no. 4, (e) subject no. 5, (f) subject no. 6, (g) subject no. 7 and (h) subject no. 8. The portions of the contigs identified at each time point are represented by the colored boxes for each subject. The relative coverage of each contig is represented, along with annotated open reading frames (ORFs) above each diagram. The length of each contig is denoted at the top of each panel.
Figure 4
Diagram of contig89 assembled from Sanger sequences from all 11 time points in subject no. 1. Putative ORFs and their direction are indicated by the arrows at the top of the diagram. ORFs that had significant homologs (BLASTP _E_-score <10−5) are indicated by the text above each arrow. Those ORFs with synteny to Rhodococcus phage Pepy6 and Poco6 (Summer et al., 2011) are labeled gp054, gp065, gp067, gp068, gp069, gp071, gp072, gp105 and gp106. The location of polymorphisms (when compared with a majority rule consensus from all time points combined) are indicated by orange vertical lines.
Figure 5
Heatmap of shared contigs across all subjects and time points. The heatmap is organized by subject and time point, where the subject number is denoted along each axis, and the individual time points are denoted by each column consecutively from Day 1 AM to Day 60 AM left to right for each subject. Each row represents an individual contig, and rows are ordered consecutively across each subject from contigs identified on Day 1 AM to Day 60 AM. The ‘matrix-like' appearance of the heatmap is due to the high intensity of homologous sequences across all time points within each subject.
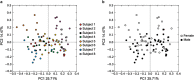
Figure 6
Principal coordinate analysis of beta diversity present in the viromes of each subject and time point. Each subject and time point are colored by subject number (a), and by gender (b).
Figure 7
Principal coordinate analysis of beta diversity present in the bacterial community measured by 16S rRNA of each subject and time point. Each subject and time point are colored by subject number (a) and gender (b).
Similar articles
- Altered oral viral ecology in association with periodontal disease.
Ly M, Abeles SR, Boehm TK, Robles-Sikisaka R, Naidu M, Santiago-Rodriguez T, Pride DT. Ly M, et al. mBio. 2014 May 20;5(3):e01133-14. doi: 10.1128/mBio.01133-14. mBio. 2014. PMID: 24846382 Free PMC article. - Evidence of a robust resident bacteriophage population revealed through analysis of the human salivary virome.
Pride DT, Salzman J, Haynes M, Rohwer F, Davis-Long C, White RA 3rd, Loomer P, Armitage GC, Relman DA. Pride DT, et al. ISME J. 2012 May;6(5):915-26. doi: 10.1038/ismej.2011.169. Epub 2011 Dec 8. ISME J. 2012. PMID: 22158393 Free PMC article. - Association between living environment and human oral viral ecology.
Robles-Sikisaka R, Ly M, Boehm T, Naidu M, Salzman J, Pride DT. Robles-Sikisaka R, et al. ISME J. 2013 Sep;7(9):1710-24. doi: 10.1038/ismej.2013.63. Epub 2013 Apr 18. ISME J. 2013. PMID: 23598790 Free PMC article. - Ecology of the Oral Microbiome: Beyond Bacteria.
Baker JL, Bor B, Agnello M, Shi W, He X. Baker JL, et al. Trends Microbiol. 2017 May;25(5):362-374. doi: 10.1016/j.tim.2016.12.012. Epub 2017 Jan 11. Trends Microbiol. 2017. PMID: 28089325 Free PMC article. Review. - Normal Oral Flora and the Oral Ecosystem.
Samaranayake L, Matsubara VH. Samaranayake L, et al. Dent Clin North Am. 2017 Apr;61(2):199-215. doi: 10.1016/j.cden.2016.11.002. Dent Clin North Am. 2017. PMID: 28317562 Review.
Cited by
- The "Gum-Gut" Axis in Inflammatory Bowel Diseases: A Hypothesis-Driven Review of Associations and Advances.
Byrd KM, Gulati AS. Byrd KM, et al. Front Immunol. 2021 Feb 19;12:620124. doi: 10.3389/fimmu.2021.620124. eCollection 2021. Front Immunol. 2021. PMID: 33679761 Free PMC article. Review. - Oral microbiome and health.
Sharma N, Bhatia S, Sodhi AS, Batra N. Sharma N, et al. AIMS Microbiol. 2018 Jan 12;4(1):42-66. doi: 10.3934/microbiol.2018.1.42. eCollection 2018. AIMS Microbiol. 2018. PMID: 31294203 Free PMC article. Review. - Deciphering the Human Virome with Single-Virus Genomics and Metagenomics.
de la Cruz Peña MJ, Martinez-Hernandez F, Garcia-Heredia I, Lluesma Gomez M, Fornas Ò, Martinez-Garcia M. de la Cruz Peña MJ, et al. Viruses. 2018 Mar 6;10(3):113. doi: 10.3390/v10030113. Viruses. 2018. PMID: 29509721 Free PMC article. - Shared and Distinct Features of Human Milk and Infant Stool Viromes.
Pannaraj PS, Ly M, Cerini C, Saavedra M, Aldrovandi GM, Saboory AA, Johnson KM, Pride DT. Pannaraj PS, et al. Front Microbiol. 2018 Jun 1;9:1162. doi: 10.3389/fmicb.2018.01162. eCollection 2018. Front Microbiol. 2018. PMID: 29910789 Free PMC article. - Effects of Long Term Antibiotic Therapy on Human Oral and Fecal Viromes.
Abeles SR, Ly M, Santiago-Rodriguez TM, Pride DT. Abeles SR, et al. PLoS One. 2015 Aug 26;10(8):e0134941. doi: 10.1371/journal.pone.0134941. eCollection 2015. PLoS One. 2015. PMID: 26309137 Free PMC article.
References
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials