Mutations in NGLY1 cause an inherited disorder of the endoplasmic reticulum-associated degradation pathway - PubMed (original) (raw)
Case Reports
doi: 10.1038/gim.2014.22. Epub 2014 Mar 20.
Vandana Shashi 2, Matthew Bainbridge 3, Michael J Gambello 4, Farah R Zahir 5, Thomas Bast 6, Rebecca Crimian 2, Kelly Schoch 2, Julia Platt 1, Rachel Cox 1, Jonathan A Bernstein 1, Mena Scavina 7, Rhonda S Walter 8, Audrey Bibb 4, Melanie Jones 4, Madhuri Hegde 4, Brett H Graham 3, Anna C Need 9, Angelica Oviedo 10, Christian P Schaaf 11, Sean Boyle 12, Atul J Butte 12, Rui Chen 13, Rong Chen 12, Michael J Clark 12, Rajini Haraksingh 12; FORGE Canada Consortium; Tina M Cowan 14, Ping He 15, Sylvie Langlois 5, Huda Y Zoghbi 16, Michael Snyder 12, Richard A Gibbs 17, Hudson H Freeze 15, David B Goldstein 18
Collaborators, Affiliations
- PMID: 24651605
- PMCID: PMC4243708
- DOI: 10.1038/gim.2014.22
Case Reports
Mutations in NGLY1 cause an inherited disorder of the endoplasmic reticulum-associated degradation pathway
Gregory M Enns et al. Genet Med. 2014 Oct.
Erratum in
- Genet Med. 2014 Jul;16(7):568. Chen, Rui [added]
Abstract
Purpose: The endoplasmic reticulum-associated degradation pathway is responsible for the translocation of misfolded proteins across the endoplasmic reticulum membrane into the cytosol for subsequent degradation by the proteasome. To define the phenotype associated with a novel inherited disorder of cytosolic endoplasmic reticulum-associated degradation pathway dysfunction, we studied a series of eight patients with deficiency of N-glycanase 1.
Methods: Whole-genome, whole-exome, or standard Sanger sequencing techniques were employed. Retrospective chart reviews were performed in order to obtain clinical data.
Results: All patients had global developmental delay, a movement disorder, and hypotonia. Other common findings included hypolacrima or alacrima (7/8), elevated liver transaminases (6/7), microcephaly (6/8), diminished reflexes (6/8), hepatocyte cytoplasmic storage material or vacuolization (5/6), and seizures (4/8). The nonsense mutation c.1201A>T (p.R401X) was the most common deleterious allele.
Conclusion: NGLY1 deficiency is a novel autosomal recessive disorder of the endoplasmic reticulum-associated degradation pathway associated with neurological dysfunction, abnormal tear production, and liver disease. The majority of patients detected to date carry a specific nonsense mutation that appears to be associated with severe disease. The phenotypic spectrum is likely to enlarge as cases with a broader range of mutations are detected.
Conflict of interest statement
Conflicts of Interest
The authors declare no conflict of interest.
Figures
Figure 1
Relative frequency of clinical findings in NGLY1 deficiency.
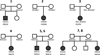
Figure 2
Pedigrees of NGLY1-deficienct patients.
Comment in
- The shifting model in clinical diagnostics: how next-generation sequencing and families are altering the way rare diseases are discovered, studied, and treated.
Might M, Wilsey M. Might M, et al. Genet Med. 2014 Oct;16(10):736-7. doi: 10.1038/gim.2014.23. Epub 2014 Mar 20. Genet Med. 2014. PMID: 24651604 No abstract available.
Similar articles
- Multi-systemic involvement in NGLY1-related disorder caused by two novel mutations.
Heeley J, Shinawi M. Heeley J, et al. Am J Med Genet A. 2015 Apr;167A(4):816-20. doi: 10.1002/ajmg.a.36889. Epub 2015 Feb 23. Am J Med Genet A. 2015. PMID: 25707956 - Ngly1 -/- rats develop neurodegenerative phenotypes and pathological abnormalities in their peripheral and central nervous systems.
Asahina M, Fujinawa R, Nakamura S, Yokoyama K, Tozawa R, Suzuki T. Asahina M, et al. Hum Mol Genet. 2020 Jun 27;29(10):1635-1647. doi: 10.1093/hmg/ddaa059. Hum Mol Genet. 2020. PMID: 32259258 Free PMC article. - Transcriptome and functional analysis in a Drosophila model of NGLY1 deficiency provides insight into therapeutic approaches.
Owings KG, Lowry JB, Bi Y, Might M, Chow CY. Owings KG, et al. Hum Mol Genet. 2018 Mar 15;27(6):1055-1066. doi: 10.1093/hmg/ddy026. Hum Mol Genet. 2018. PMID: 29346549 Free PMC article. - Novel NGLY1 gene variants in Chinese children with global developmental delay, microcephaly, hypotonia, hypertransaminasemia, alacrimia, and feeding difficulty.
Abuduxikuer K, Zou L, Wang L, Chen L, Wang JS. Abuduxikuer K, et al. J Hum Genet. 2020 Apr;65(4):387-396. doi: 10.1038/s10038-019-0719-9. Epub 2020 Jan 21. J Hum Genet. 2020. PMID: 31965062 Review. - The cytoplasmic peptide:N-glycanase (NGLY1) - Structure, expression and cellular functions.
Suzuki T, Huang C, Fujihira H. Suzuki T, et al. Gene. 2016 Feb 10;577(1):1-7. doi: 10.1016/j.gene.2015.11.021. Epub 2015 Nov 30. Gene. 2016. PMID: 26611529 Free PMC article. Review.
Cited by
- Generation and degradation of free asparagine-linked glycans.
Harada Y, Hirayama H, Suzuki T. Harada Y, et al. Cell Mol Life Sci. 2015 Jul;72(13):2509-33. doi: 10.1007/s00018-015-1881-7. Epub 2015 Mar 14. Cell Mol Life Sci. 2015. PMID: 25772500 Free PMC article. Review. - Gut barrier defects, intestinal immune hyperactivation and enhanced lipid catabolism drive lethality in NGLY1-deficient Drosophila.
Pandey A, Galeone A, Han SY, Story BA, Consonni G, Mueller WF, Steinmetz LM, Vaccari T, Jafar-Nejad H. Pandey A, et al. Nat Commun. 2023 Sep 13;14(1):5667. doi: 10.1038/s41467-023-40910-w. Nat Commun. 2023. PMID: 37704604 Free PMC article. - Sugar-Recognizing Ubiquitin Ligases: Action Mechanisms and Physiology.
Yoshida Y, Mizushima T, Tanaka K. Yoshida Y, et al. Front Physiol. 2019 Feb 19;10:104. doi: 10.3389/fphys.2019.00104. eCollection 2019. Front Physiol. 2019. PMID: 30837888 Free PMC article. Review. - Data sharing in the undiagnosed diseases network.
Brownstein CA, Holm IA, Ramoni R, Goldstein DB; Members of the Undiagnosed Diseases Network. Brownstein CA, et al. Hum Mutat. 2015 Oct;36(10):985-8. doi: 10.1002/humu.22840. Epub 2015 Aug 27. Hum Mutat. 2015. PMID: 26220576 Free PMC article. - Prospective phenotyping of NGLY1-CDDG, the first congenital disorder of deglycosylation.
Lam C, Ferreira C, Krasnewich D, Toro C, Latham L, Zein WM, Lehky T, Brewer C, Baker EH, Thurm A, Farmer CA, Rosenzweig SD, Lyons JJ, Schreiber JM, Gropman A, Lingala S, Ghany MG, Solomon B, Macnamara E, Davids M, Stratakis CA, Kimonis V, Gahl WA, Wolfe L. Lam C, et al. Genet Med. 2017 Feb;19(2):160-168. doi: 10.1038/gim.2016.75. Epub 2016 Jul 7. Genet Med. 2017. PMID: 27388694 Free PMC article.
References
- Takahashi N. Demonstration of a new amidase acting on glycopeptides. Biochem Biophys Res Commun. 1977;76:1194–1201. - PubMed
- Plummer TH, Jr, Elder JH, Alexander S, Phelan AW, Tarentino AL. Demonstration of peptide:N-glycosidase F activity in endo-beta-N-acetylglucosaminidase F preparations. J Biol Chem. 1984;259:10700–10704. - PubMed
- Xin F, Wang S, Song L, Liang Q, Qi Q. Molecular identification and characterization of peptide: N-glycanase from Schizosaccharomyces pombe. Biochem Biophys Res Commun. 2008;368:907–912. - PubMed
- Masahara-Negishi Y, Hosomi A, Della Mea M, Serafini-Fracassini D, Suzuki T. A plant peptide: N-glycanase orthologue facilitates glycoprotein ER-associated degradation in yeast. Biochim Biophys Acta. 2012;1820:1457–1462. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U54-HG003273/HG/NHGRI NIH HHS/United States
- CAPMC/ CIHR/Canada
- U54 HD083092/HD/NICHD NIH HHS/United States
- U54 HG003273/HG/NHGRI NIH HHS/United States
- T32 HG000044/HG/NHGRI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials