Erythro-megakaryocytic transcription factors associated with hereditary anemia - PubMed (original) (raw)
Review
Erythro-megakaryocytic transcription factors associated with hereditary anemia
John D Crispino et al. Blood. 2014.
Abstract
Most heritable anemias are caused by mutations in genes encoding globins, red blood cell (RBC) membrane proteins, or enzymes in the glycolytic and hexose monophosphate shunt pathways. A less common class of genetic anemia is caused by mutations that alter the functions of erythroid transcription factors (TFs). Many TF mutations associated with heritable anemia cause truncations or amino acid substitutions, resulting in the production of functionally altered proteins. Characterization of these mutant proteins has provided insights into mechanisms of gene expression, hematopoietic development, and human disease. Mutations within promoter or enhancer regions that disrupt TF binding to essential erythroid genes also cause anemia and heritable variations in RBC traits, such as fetal hemoglobin content. Defining the latter may have important clinical implications for de-repressing fetal hemoglobin synthesis to treat sickle cell anemia and β thalassemia. Functionally important alterations in genes encoding TFs or their cognate cis elements are likely to occur more frequently than currently appreciated, a hypothesis that will soon be tested through ongoing genome-wide association studies and the rapidly expanding use of global genome sequencing for human diagnostics. Findings obtained through such studies of RBCs and associated diseases are likely generalizable to many human diseases and quantitative traits.
© 2014 by The American Society of Hematology.
Figures
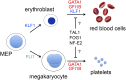
Figure 1
TFs that drive erythromegakaryopoiesis. RBCs and MEGs derive from the bipotential MEP. KLF1 and FLI1 regulate lineage determination of the MEP and subsequent erythroid or MEG maturation, respectively. GATA1, TAL1, FOG1, NF-E2, and GFI1B regulate the maturation of both lineages. Germ-line mutations in KLF1 (shown in blue) cause isolated congenital anemia, whereas mutations in the TFs indicated in red affect RBCs and MEGs. Candidate TFs for which no germ-line human mutations have yet been discovered to be associated with anemia or thrombocytopenia are shown in black. Deficiency of FLI1 (shown in green) is associated with thrombocytopenia, but not anemia in Paris-Trousseau syndrome.
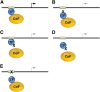
Figure 2
Mutations affecting transcription factor function. (A) TFs and bound cofactors (Cofs) regulate gene expression by binding cognate cis elements within DNA regulatory regions including promoters or enhancers (gray box). The arrow indicates gene transcription. (B) Mutations in the TF DNA binding domain inhibit TF interactions with some or all cis elements. The dashed lines indicate altered transcription. (C) TF mutations that inhibit Cof binding alter gene expression at some or all target genes. (D) Some Cof interactions are required for in vivo DNA occupancy by TFs. Thus, TF mutations outside of the DNA binding domain can inhibit gene occupancy. (E) Mutations in DNA cis elements can impair TF occupancy at a single site within one gene, thereby altering its transcription.
Figure 3
GATA1 and KLF1 domain modules, mutations, and associated diseases. (A) (Top) Three domains of GATA1 are shown: the N and C-terminal zinc fingers (Nf and Cf) and the NAD. (Middle) Mutations in the Nf (noted by asterisks) disrupt DNA binding or associations with the essential cofactors Friend of GATA1 (FOG-1) or TAL1 complex. These germ-line mutations cause a variety of inherited anemias and/or thrombocytopenias. (Bottom) Distinct mutations within exon 2 lead to predominant expression of GATA1s, which retains both zinc fingers, but is missing the NAD. In infants and young children with DS (trisomy 21), somatic mutations that favor GATA1s production cause TMD and AMKL. In euploid patients, germ-line GATA1s mutations are associated with congenital anemia, including DBA. (B) KLF1 includes an N-terminal proline-rich domain and 3 C-terminal zinc finger domains. Various missense and frameshift mutations (noted by asterisks) throughout the gene are associated with hereditary anemias, altered Lutheran blood group expression, and/or persistence of fetal hemoglobin expression. Note that E325K has a dominant negative effect. Adapted from Singleton et al.
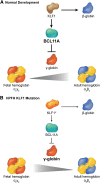
Figure 4
Interplay between KLF1 and BCL11a in the regulation of hemoglobin switching. (A) During normal development, KLF1 activates transcription of the BCL11A gene, which in turn represses γ globin expression promoting the switch from fetal (α2γ2) to adult (α2β2) hemoglobin. Simultaneously, high levels of KLF1 activate β-globin gene expression. (B) In some cases of HPFH, haplo-insufficiency of KLF1 (KLF1*) is associated with reduced BCL11A expression, which allows for persistence of fetal hemoglobin. Simultaneously, reduced KLF1 fails to activate β-globin expression.
Similar articles
- Genomic footprinting and sequencing of human beta-globin locus. Tissue specificity and cell line artifact.
Reddy PM, Stamatoyannopoulos G, Papayannopoulou T, Shen CK. Reddy PM, et al. J Biol Chem. 1994 Mar 18;269(11):8287-95. J Biol Chem. 1994. PMID: 8132552 - Characterization of a functional ZBP-89 binding site that mediates Gata1 gene expression during hematopoietic development.
Ohneda K, Ohmori S, Ishijima Y, Nakano M, Yamamoto M. Ohneda K, et al. J Biol Chem. 2009 Oct 30;284(44):30187-99. doi: 10.1074/jbc.M109.026948. Epub 2009 Sep 1. J Biol Chem. 2009. PMID: 19723625 Free PMC article. - Ablation of Nrf2 function does not increase the erythroid or megakaryocytic cell lineage dysfunction caused by p45 NF-E2 gene disruption.
Kuroha T, Takahashi S, Komeno T, Itoh K, Nagasawa T, Yamamoto M. Kuroha T, et al. J Biochem. 1998 Mar;123(3):376-9. doi: 10.1093/oxfordjournals.jbchem.a021947. J Biochem. 1998. PMID: 9538217 - GATA transcription factors in hematologic disease.
Cantor AB. Cantor AB. Int J Hematol. 2005 Jun;81(5):378-84. doi: 10.1532/ijh97.04180. Int J Hematol. 2005. PMID: 16158817 Review. - Severe anemia caused by dominant mutations in Krüppel-like factor 1 (KLF1).
Kulczynska-Figurny K, Bieker JJ, Siatecka M. Kulczynska-Figurny K, et al. Mutat Res Rev Mutat Res. 2020 Oct-Dec;786:108336. doi: 10.1016/j.mrrev.2020.108336. Epub 2020 Oct 9. Mutat Res Rev Mutat Res. 2020. PMID: 33339573 Free PMC article. Review.
Cited by
- Acute Megakaryocytic Leukemia.
McNulty M, Crispino JD. McNulty M, et al. Cold Spring Harb Perspect Med. 2020 Feb 3;10(2):a034884. doi: 10.1101/cshperspect.a034884. Cold Spring Harb Perspect Med. 2020. PMID: 31548219 Free PMC article. Review. - A novel 33-Gene targeted resequencing panel provides accurate, clinical-grade diagnosis and improves patient management for rare inherited anaemias.
Roy NB, Wilson EA, Henderson S, Wray K, Babbs C, Okoli S, Atoyebi W, Mixon A, Cahill MR, Carey P, Cullis J, Curtin J, Dreau H, Ferguson DJ, Gibson B, Hall G, Mason J, Morgan M, Proven M, Qureshi A, Sanchez Garcia J, Sirachainan N, Teo J, Tedgård U, Higgs D, Roberts D, Roberts I, Schuh A. Roy NB, et al. Br J Haematol. 2016 Oct;175(2):318-330. doi: 10.1111/bjh.14221. Epub 2016 Jul 19. Br J Haematol. 2016. PMID: 27432187 Free PMC article. - A Positive Regulatory Feedback Loop between EKLF/KLF1 and TAL1/SCL Sustaining the Erythropoiesis.
Hung CH, Lee TL, Huang AY, Yang KC, Shyu YC, Wen SC, Lu MJ, Yuan S, Shen CJ. Hung CH, et al. Int J Mol Sci. 2021 Jul 27;22(15):8024. doi: 10.3390/ijms22158024. Int J Mol Sci. 2021. PMID: 34360789 Free PMC article. - Cytopenia: a report of haplo-cord transplantation in twin brothers caused by a novel germline GATA1 mutation and family survey.
Sun XH, Liu Q, Wu SN, Xu WH, Chen K, Shao JB, Jiang H. Sun XH, et al. Ann Hematol. 2023 Nov;102(11):3177-3184. doi: 10.1007/s00277-023-05363-7. Epub 2023 Jul 18. Ann Hematol. 2023. PMID: 37460606 - Inhibition of human primary megakaryocyte differentiation by anagrelide: a gene expression profiling analysis.
Sakurai K, Fujiwara T, Hasegawa S, Okitsu Y, Fukuhara N, Onishi Y, Yamada-Fujiwara M, Ichinohasama R, Harigae H. Sakurai K, et al. Int J Hematol. 2016 Aug;104(2):190-9. doi: 10.1007/s12185-016-2006-2. Epub 2016 Apr 15. Int J Hematol. 2016. PMID: 27084257
References
- Bouilloux F, Juban G, Cohet N, et al. EKLF restricts megakaryocytic differentiation at the benefit of erythrocytic differentiation. Blood. 2008;112(3):576–584. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01DK092318/DK/NIDDK NIH HHS/United States
- R01 DK092318/DK/NIDDK NIH HHS/United States
- R01 DK065806/DK/NIDDK NIH HHS/United States
- R01DK065806/DK/NIDDK NIH HHS/United States
- R01 DK101329/DK/NIDDK NIH HHS/United States
- P30 DK090969/DK/NIDDK NIH HHS/United States
- P30DK090969/DK/NIDDK NIH HHS/United States
- DK101329/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous