Novel betacoronavirus in dromedaries of the Middle East, 2013 - PubMed (original) (raw)
doi: 10.3201/eid2004.131769.
Susanna K P Lau, Ulrich Wernery, Emily Y M Wong, Alan K L Tsang, Bobby Johnson, Cyril C Y Yip, Candy C Y Lau, Saritha Sivakumar, Jian-Piao Cai, Rachel Y Y Fan, Kwok-Hung Chan, Ringu Mareena, Kwok-Yung Yuen
- PMID: 24655427
- PMCID: PMC3966378
- DOI: 10.3201/eid2004.131769
Novel betacoronavirus in dromedaries of the Middle East, 2013
Patrick C Y Woo et al. Emerg Infect Dis. 2014 Apr.
Abstract
In 2013, a novel betacoronavirus was identified in fecal samples from dromedaries in Dubai, United Arab Emirates. Antibodies against the recombinant nucleocapsid protein of the virus, which we named dromedary camel coronavirus (DcCoV) UAE-HKU23, were detected in 52% of 59 dromedary serum samples tested. In an analysis of 3 complete DcCoV UAE-HKU23 genomes, we identified the virus as a betacoronavirus in lineage A1. The DcCoV UAE-HKU23 genome has G+C contents; a general preference for G/C in the third position of codons; a cleavage site for spike protein; and a membrane protein of similar length to that of other betacoronavirus A1 members, to which DcCoV UAE-HKU23 is phylogenetically closely related. Along with this coronavirus, viruses of at least 8 other families have been found to infect camels. Because camels have a close association with humans, continuous surveillance should be conducted to understand the potential for virus emergence in camels and for virus transmission to humans.
Keywords: DcCoV; DcCoVUAE-HKU23; Dubai; MERS-CoV; Middle East; Middle East respiratory syndrome coronavirus; United Arab Emirates; betacoronavirus; camel; camel coronavirus; coronavirus; dromedaries; dromedary; dromedary camel coronavirus UAE-HKU23; dromedary coronavirus; viruses; zoonoses.
Figures
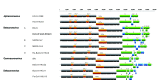
Figure 1
Genome organizations of a novel betacoronavirus, in boldface, discovered in dromedaries in the Middle East in 2013, and representative coronaviruses from each coronavirus genus (labeled on left). Numbers at top represent genome position. A, B, C, and D represent betacoronavirus lineages. Papain-like proteases (PL1pro, PL2pro, and PLpro), chymotrypsin-like protease (3CLpro), and RNA-dependent RNA polymerase (RdRp) are represented by orange boxes. Hemagglutinin esterase (HE) and spike (S), envelope (E), membrane (M), and nucleocapsid (N) proteins are represented by green boxes. Putative accessory proteins are represented by blue boxes. HCoV, human coronavirus; BCoV, bovine CoV; DcCoV, dromedary camel CoV; SARS-CoV, severe acute respiratory syndrome CoV; MERS-CoV, Middle East respiratory syndrome CoV; Ro-BatCoV, rousettus bat CoV; IBV, infectious bronchitis virus; BdCoV, bottlenose dolphin CoV; BuCoV, bulbul CoV; PorCoV, porcine CoV.
Figure 2
Amino acid comparison of the spike protein of a novel betacoronavirus, dromedary camel coronavirus (DcCoV) UAE-HKU23, discovered in dromedaries in the Middle East in 2013, with that of bovine coronavirus (BCoV; GenBank accession no. AF391541).Amino acid substitution sites, key amino acids for virulence and receptor binding in BCoV, and cleavage sites are shown. Amino acid positions are given with reference to DcCoV UAE-HKU23. Conserved amino acids, compared with those of DcCoV UAE-HKU23 (strain 265F), are represented by dots. Amino acids of putative cleavage sites are underlined. Amino acids within the S1 hypervariable region of BCoV are marked with open boxes. Amino acid sites central to virulence in BCoV are highlighted in light gray. Amino acid sites shown to affect S1-mediated receptor binding in BCoV are highlighted in dark gray. The 4 critical sugar-binding residues are indicated by black dots. The amino acid site that discriminated between enteric and respiratory BCoV strains is highlighted in black.
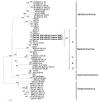
Figure 3
Phylogenetic analysis of open reading frame (ORF) 1b polyprotein of dromedary camel coronavirus (DcCoV) UAE-HKU23 from dromedaries of the Middle East, 2013. The tree was constructed by the neighbor-joining method, using Jones-Taylor-Thornton substitution model with gamma distributed rate variation and bootstrap values calculated from 1,000 trees. Bootstrap values of <70% are not shown. A total of 2,725 aa positions in ORF1b polyprotein were included in the analysis. The tree was rooted to Breda virus (GenBank accession no. AY_427798). Betacoronavirus lineages A1 and A�?"D are indicated on the right. Boldface indicates the 3 strains of DcCoV UAE-HKU23 characterized in this study. Virus definitions and GenBank accession numbers (in parentheses) follow: Mi-BatCoV 1B, miniopterus bat CoV 1B (NC_010436); Mi-BatCoV 1A (NC_010437); Mi-BatCoV HKU8 (NC_010438); Ro-BatCoV HKU10, rousettus bat CoV HKU10 (JQ989270); Hi-BatCoV HKU10, hipposideros bat CoV HKU10 (JQ989266); HCoV-NL63, human CoV NL63 (NC_005831); HCoV-229E (NC_002645); RhBatCoV HKU2, rhinolophus bat CoV HKU2 (EF203064); Sc-BatCoV-512, scotophilus bat CoV 512 (NC_009657); PEDV, porcine epidemic diarrhea virus (NC_003436);MCoV, mink CoV (HM245925);CCoV, canine CoV (GQ477367); FIPV, feline infectious peritonitis virus (AY994055); PRCV, porcine respiratory CoV; TGEV, transmissible gastroenteritis virus; SACoV, sable antelope CoV; GiCoV, giraffe CoV (EF424622); CRCoV, canine respiratory CoV (JX860640); BCoV, bovine CoV (NC_003045); HCoV-OC43, human CoV OC43 (NC_005147);PHEV, porcine hemagglutinating encephalomyelitis virus (NC_007732); ECoV, equine CoV (NC_010327); RbCoV HKU14, rabbit CoV HKU14 (JN874559); HCoV-HKU1, human CoV HKU1 (NC_006577); MHV, murine hepatitis virus (NC_001846); RCoV, rat CoV (NC_012936); Pi-BatCoV HKU5, pipistrellus bat CoV HKU5 (NC_009020); MERS-CoV, Middle East respiratory syndrome CoV (JX869059);Ty-BatCoV HKU4, tylonycteris bat CoV HKU4 (NC_009019); SARSr-CiCoV, SARS-related palm civet CoV (AY304488); SARS-CoV, severe acute respiratory syndrome�?"associated human CoV (NC_004718); SARSrCoV CFB, SARS-related Chinese ferret badger CoV (AY545919); SARS-related rhinolophus bat CoV HKU3 (DQ022305); Ro-BatCoV HKU9, rousettus bat CoV HKU9 (NC_009021); TCoV, turkey CoV (NC_010800); IBV-peafowl, peafowl CoV (AY641576); IBV, infectious bronchitis virus (NC_001451); IBV-partridge, partridge CoV (AY646283); BdCoV HKU22, bottlenose dolphin CoV HKU22 (KF793824); BWCoV-SW1, Beluga whale CoV SW1 (NC_010646); WiCoV HKU20, wigeon CoV HKU20 (JQ065048); NHCoV HKU19, night-heron CoV HKU19 (JQ065047); CMCoV HKU21, common-moorhen CoV HKU21 (JQ065049); BuCoV HKU11, bulbul CoV HKU11 (FJ376619); ThCoV HKU12, thrush CoV HKU12 (FJ376621); WECoV HKU16, white-eye CoV HKU16 (JQ065044); MunCoV HKU13, munia CoV HKU13 (FJ376622); MRCoV HKU18, magpie�?"robin CoV HKU18 (JQ065046); PorCoV HKU15, porcine CoV HKU15 (JQ065042); SpCoV HKU17, sparrow CoV HKU17 (JQ065045). Numbers at nodes represent bootstrap values. Scale bar indicates the estimated number of substitutions per 10 aa.
Figure 4
Phylogenetic analyses of spike protein of dromedary camel coronavirus (DcCoV) UAE-HKU23from dromedaries of the Middle East, 2013.The tree was constructed by the neighbor-joining method, using Jones-Taylor-Thornton substitution model with gamma distributed rate variation and bootstrap values calculated from 1,000 trees. Bootstrap values of <70% are not shown. A total of 1,366 aa positions in spike protein were included in the analysis. The tree was rooted to Breda virus (GenBank accession no. AY_427798).Betacoronavirus lineages A1 and A�?"D are indicated on the right. Boldface indicates the 3 strains of DcCoV UAE-HKU23 characterized in this study. Virus definitions and GenBank accession numbers (in parentheses) follow: RbCoV HKU14, rabbit CoV HKU14 (JN874559); HCoV-OC43, human CoV OC43 (NC_005147); CRCoV, canine respiratory CoV (JX860640); BCoV, bovine CoV (NC_003045); SACoV, sable antelope CoV (EF424621); GiCoV, giraffe CoV (EF424622); PHEV, porcine hemagglutinating encephalomyelitis virus (NC_007732); ECoV, equine CoV (NC_010327); MHV, murine hepatitis virus (NC_001846); RCoV, rat CoV (NC_012936); HCoV-HKU1, human CoV HKU1 (NC_006577); MERS-CoV, Middle East respiratory syndrome CoV (JX869059); Ty-BatCoV HKU4, tylonycteris bat CoV HKU4 (NC_009019); Pi-BatCoV HKU5, pipistrellus bat CoV HKU5 (NC_009020); SARSrCoV CFB, SARS-related Chinese ferret badger CoV (AY545919); SARSr-CiCoV, SARS-related palm civet CoV (AY304488); SARS-CoV, severe acute respiratory syndrome�?"associated human CoV (NC_004718); SARSr-Rh-BatCoV HKU3, SARS-related rhinolophus bat CoV HKU3 (DQ022305); Ro-BatCoV HKU9, rousettus bat CoV HKU9 (NC_009021); RhBatCoV HKU2, rhinolophus bat CoV HKU2 (EF203064); BWCoV-SW1, Beluga whale CoV SW1 (NC_010646); BdCoV HKU22, bottlenose dolphin CoV HKU22 (KF793824); TCoV, turkey CoV (NC_010800); IBV-partridge, partridge CoV (AY646283); IBV, infectious bronchitis virus (NC_001451); IBV-peafowl, peafowl CoV (AY641576); CCoV, canine CoV (GQ477367); FIPV, feline infectious peritonitis virus (AY994055); PRCV, porcine respiratory CoV (DQ811787); TGEV, transmissible gastroenteritis virus (DQ811789); MCoV, mink CoV (HM245925); Sc-BatCoV-512, scotophilus bat CoV 512 (NC_009657); PEDV, porcine epidemic diarrhea virus (NC_003436); Mi-BatCoV 1B, miniopterus bat CoV 1B (NC_010436); Mi-BatCoV 1A, miniopterus bat CoV 1A (NC_010437); Mi-BatCoV HKU8, miniopterus bat CoV HKU8 (NC_010438); Ro-BatCoV HKU10, rousettus bat CoV HKU10 (JQ989270); Hi-BatCoV HKU10, hipposideros bat CoV HKU10 (JQ989266); HCoV-NL63, human CoV NL63 (NC_005831); HCoV-229E, human CoV 229E (NC_002645); WiCoV HKU20, wigeon CoV HKU20 (JQ065048); MRCoV HKU18, magpie�?"robin CoV HKU18 (JQ065046); SpCoV HKU17, sparrow CoV HKU17 (JQ065045); NHCoV HKU19, night-heron CoV HKU19 (JQ065047); ThCoV HKU12, thrush CoV HKU12 (FJ376621); CMCoV HKU21, common-moorhen CoV HKU21 (JQ065049); WECoV HKU16, white-eye CoV HKU16 (JQ065044); MunCoV HKU13, munia CoV HKU13 (FJ376622); PorCoV HKU15, porcine CoV HKU15 (JQ065042); BuCoV HKU11, bulbul CoV HKU11 (FJ376619). Numbers at nodes represent bootstrap values. Scale bar indicates the estimated number of substitutions per 5 aa.
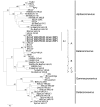
Figure 5
Phylogenetic analyses of the nucleocapsid protein of a novel coronavirus (CoV), dromedary camel CoV (DcCoV) UAE-HKU23, discovered in dromedaries of the Middle East, 2013.The tree was constructed by the neighbor-joining method, using Jones-Taylor-Thornton substitution model with gamma distributed rate variation and bootstrap values calculated from 1,000 trees. Bootstrap values of <70% are not shown. A total of 448 aa positions were included in the analysis. The tree was rooted to Breda virus (GenBank accession no. AY_427798). Betacoronavirus lineages A1 and A�?"D are indicated on the right. Boldface indicates the 3 strains of DcCoV UAE-HKU23 characterized in this study. Virus definitions and GenBank accession numbers (in parentheses) follow: TGEV, transmissible gastroenteritis virus (DQ811789); PRCV, porcine respiratory CoV (DQ811787); CCoV, canine CoV (GQ477367); FIPV, feline infectious peritonitis virus (AY994055); MCoV, mink CoV (HM245925); RhBatCoV HKU2, rhinolophus bat CoV HKU2 (EF203064); Mi-BatCoV 1A, miniopterus bat CoV 1A (NC_010437); Mi-BatCoV 1B, miniopterus bat CoV 1B (NC_010436); Mi-BatCoV HKU8, miniopterus bat CoV HKU8 (NC_010438); HCoV-229E, human CoV 229E (NC_002645); PEDV, porcine epidemic diarrhea virus (NC_003436); Sc-BatCoV-512, scotophilus bat CoV 512 (NC_009657); HCoV-NL63, human CoV NL63 (NC_005831); Hi-BatCoV HKU10, hipposideros bat CoV HKU10 (JQ989266); Ro-BatCoV HKU10, rousettus bat CoV HKU10 (JQ989270); RbCoV HKU14, rabbit CoV HKU14 (JN874559); HCoV-OC43, human CoV OC43 (NC_005147); PHEV, porcine hemagglutinating encephalomyelitis virus (NC_007732); CRCoV, canine respiratory CoV (JX860640); BCoV, bovine CoV (NC_003045); GiCoV, giraffe CoV (EF424622); SACoV, sable antelope CoV (EF424621); ECoV, equine CoV (NC_010327); HCoV-HKU1, human CoV HKU1 (NC_006577); MHV, murine hepatitis virus (NC_001846); RCoV, rat CoV (NC_012936); SARSr-CiCoV, SARS-related palm civet CoV (AY304488); SARS-CoV, severe acute respiratory syndrome�?"associated human CoV (NC_004718); SARSrCoV CFB, SARS-related Chinese ferret badger CoV (AY545919); SARSr-Rh-BatCoV HKU3, SARS-related rhinolophus bat CoV HKU3 (DQ022305); MERS-CoV, Middle East respiratory syndrome CoV (JX869059); Pi-BatCoV HKU5, pipistrellus bat CoV HKU5 (NC_009020); Ty-BatCoV HKU4, tylonycteris bat CoV HKU4 (NC_009019); Ro-BatCoV HKU9, rousettus bat CoV HKU9 (NC_009021); IBV, infectious bronchitis virus (NC_001451); IBV-partridge, partridge CoV (AY646283); TCoV, turkey CoV (NC_010800); IBV-peafowl, peafowl CoV (AY641576); BWCoV-SW1, Beluga whale CoV SW1 (NC_010646); BdCoV HKU22, bottlenose dolphin CoV HKU22 (KF793824); WiCoV HKU20, wigeon CoV HKU20 (JQ065048); CMCoV HKU21, common-moorhen CoV HKU21 (JQ065049); NHCoV HKU19, night-heron CoV HKU19 (JQ065047); BuCoV HKU11, bulbul CoV HKU11 (FJ376619); MunCoV HKU13, munia CoV HKU13 (FJ376622); MRCoV HKU18, magpie�?"robin CoV HKU18 (JQ065046); ThCoV HKU12, thrush CoV HKU12 (FJ376621); WECoV HKU16, white-eye CoV HKU16 (JQ065044); PorCoV HKU15, porcine CoV HKU15 (JQ065042); SpCoV HKU17, sparrow CoV HKU17 (JQ065045). Numbers at nodes represent bootstrap values. Scale bar indicates the estimated number of substitutions per 5 aa.
Figure 6
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blot analysis of a novel coronavirus, dromedary camel coronavirus UAE-HKU23, discovered in dromedaries of the Middle East, 2013.Nucleocapsid protein was expressed in Escherichia coli. M, protein molecular-mass marker; kDa, kilodaltons. Lanes: 1, non-induced crude E. coli cell lysate; 2, induced crude E. coli cell lysate of DcCoV UAE-HKU23 nucleocapsid protein; 3, purified recombinant DcCoV UAE-HKU23 nucleocapsid protein; 4, dromedary camel serum sample strongly positive for antibody against nucleocapsid protein; 5, dromedary camel serum sample moderately positive for antibody against nucleocapsid protein; 6 and 7: dromedary camel serum sample negative for antibody against nucleocapsid protein.
Figure 7
The evolution of corona viruses from their ancestors in bat and bird hosts to new virus species that infect other animals.
Similar articles
- Isolation and Characterization of Dromedary Camel Coronavirus UAE-HKU23 from Dromedaries of the Middle East: Minimal Serological Cross-Reactivity between MERS Coronavirus and Dromedary Camel Coronavirus UAE-HKU23.
Woo PC, Lau SK, Fan RY, Lau CC, Wong EY, Joseph S, Tsang AK, Wernery R, Yip CC, Tsang CC, Wernery U, Yuen KY. Woo PC, et al. Int J Mol Sci. 2016 May 7;17(5):691. doi: 10.3390/ijms17050691. Int J Mol Sci. 2016. PMID: 27164099 Free PMC article. - Diversity of Dromedary Camel Coronavirus HKU23 in African Camels Revealed Multiple Recombination Events among Closely Related Betacoronaviruses of the Subgenus Embecovirus.
So RTY, Chu DKW, Miguel E, Perera RAPM, Oladipo JO, Fassi-Fihri O, Aylet G, Ko RLW, Zhou Z, Cheng MS, Kuranga SA, Roger FL, Chevalier V, Webby RJ, Woo PCY, Poon LLM, Peiris M. So RTY, et al. J Virol. 2019 Nov 13;93(23):e01236-19. doi: 10.1128/JVI.01236-19. Print 2019 Dec 1. J Virol. 2019. PMID: 31534035 Free PMC article. - Antibodies against MERS coronavirus in dromedary camels, United Arab Emirates, 2003 and 2013.
Meyer B, Müller MA, Corman VM, Reusken CB, Ritz D, Godeke GJ, Lattwein E, Kallies S, Siemens A, van Beek J, Drexler JF, Muth D, Bosch BJ, Wernery U, Koopmans MP, Wernery R, Drosten C. Meyer B, et al. Emerg Infect Dis. 2014 Apr;20(4):552-9. doi: 10.3201/eid2004.131746. Emerg Infect Dis. 2014. PMID: 24655412 Free PMC article. - Middle East respiratory syndrome coronavirus antibody reactors among camels in Dubai, United Arab Emirates, in 2005.
Alexandersen S, Kobinger GP, Soule G, Wernery U. Alexandersen S, et al. Transbound Emerg Dis. 2014 Apr;61(2):105-8. doi: 10.1111/tbed.12212. Epub 2014 Jan 24. Transbound Emerg Dis. 2014. PMID: 24456414 Free PMC article. Review. - Middle East respiratory syndrome (MERS) coronavirus and dromedaries.
Wernery U, Lau SK, Woo PC. Wernery U, et al. Vet J. 2017 Feb;220:75-79. doi: 10.1016/j.tvjl.2016.12.020. Epub 2017 Jan 9. Vet J. 2017. PMID: 28190501 Free PMC article. Review.
Cited by
- A systematic approach to novel virus discovery in emerging infectious disease outbreaks.
Sridhar S, To KK, Chan JF, Lau SK, Woo PC, Yuen KY. Sridhar S, et al. J Mol Diagn. 2015 May;17(3):230-41. doi: 10.1016/j.jmoldx.2014.12.002. Epub 2015 Mar 4. J Mol Diagn. 2015. PMID: 25746799 Free PMC article. Review. - Crystallographic analysis of the N-terminal domain of Middle East respiratory syndrome coronavirus nucleocapsid protein.
Wang YS, Chang CK, Hou MH. Wang YS, et al. Acta Crystallogr F Struct Biol Commun. 2015 Aug;71(Pt 8):977-80. doi: 10.1107/S2053230X15010146. Epub 2015 Jul 28. Acta Crystallogr F Struct Biol Commun. 2015. PMID: 26249685 Free PMC article. - Middle East Respiratory Syndrome Coronavirus Antibodies in Bactrian and Hybrid Camels from Dubai.
Lau SKP, Li KSM, Luk HKH, He Z, Teng JLL, Yuen KY, Wernery U, Woo PCY. Lau SKP, et al. mSphere. 2020 Jan 22;5(1):e00898-19. doi: 10.1128/mSphere.00898-19. mSphere. 2020. PMID: 31969478 Free PMC article. - A scenario-based evaluation of the Middle East respiratory syndrome coronavirus and the Hajj.
Gardner LM, Rey D, Heywood AE, Toms R, Wood J, Travis Waller S, Raina MacIntyre C. Gardner LM, et al. Risk Anal. 2014 Aug;34(8):1391-400. doi: 10.1111/risa.12253. Epub 2014 Jul 14. Risk Anal. 2014. PMID: 25041625 Free PMC article. - First detection of equine coronavirus (ECoV) in Europe.
Miszczak F, Tesson V, Kin N, Dina J, Balasuriya UB, Pronost S, Vabret A. Miszczak F, et al. Vet Microbiol. 2014 Jun 25;171(1-2):206-9. doi: 10.1016/j.vetmic.2014.03.031. Epub 2014 Mar 30. Vet Microbiol. 2014. PMID: 24768449 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous