High rates of antimicrobial drug resistance gene acquisition after international travel, The Netherlands - PubMed (original) (raw)
High rates of antimicrobial drug resistance gene acquisition after international travel, The Netherlands
Christian J H von Wintersdorff et al. Emerg Infect Dis. 2014 Apr.
Abstract
We investigated the effect of international travel on the gut resistome of 122 healthy travelers from the Netherlands by using a targeted metagenomic approach. Our results confirm high acquisition rates of the extended-spectrum β-lactamase encoding gene blaCTX-M, documenting a rise in prevalence from 9.0% before travel to 33.6% after travel (p<0.001). The prevalence of quinolone resistance encoding genes qnrB and qnrS increased from 6.6% and 8.2% before travel to 36.9% and 55.7% after travel, respectively (both p<0.001). Travel to Southeast Asia and the Indian subcontinent was associated with the highest acquisition rates of qnrS and both blaCTX-M and qnrS, respectively. Investigation of the associations between the acquisitions of the blaCTX-M and qnr genes showed that acquisition of a blaCTX-M gene was not associated with that of a qnrB (p = 0.305) or qnrS (p = 0.080) gene. These findings support the increasing evidence that travelers contribute to the spread of antimicrobial drug resistance.
Keywords: CTX-M; ESBL; Enterobacteriaceae; Kluyvera; Shewanella algae; antibacterial; antibiotics; antimicrobial; bacteria; intestinal microbiota; metagenomic; qnrB; qnrS; quinolone; resistance genes; the Netherlands; traveling.
Figures
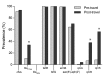
Figure 1
Prevalence (%) of antimicrobial drug resistance determinants in fecal samples from 122 healthy travelers from the Netherlands before and after travel, 2010–2012. Statistical significance of the prevalence between the 2 groups was calculated by using the McNemar test for paired samples and is indicated by * (p<0.001).
Figure 2
Relative changes in gene abundance before and after travel for each of 122 healthy travelers from the Netherlands during 2010–2012 for genes cfxA (A), tetM (B), tetQ (C), and ermB (D). Increases are shown with white bars on the positive _y_-axis; decreases are shown in dark gray bars on the negative _y_-axis. Each bar on the _x_-axis represents the change in a different study participant. The travel destination regions of the participants are indicated above the graph. No region is indicated for some travelers who either visited >1 of these regions or visited countries that were not in the defined regions (see Table 2).
Similar articles
- Destination shapes antibiotic resistance gene acquisitions, abundance increases, and diversity changes in Dutch travelers.
D'Souza AW, Boolchandani M, Patel S, Galazzo G, van Hattem JM, Arcilla MS, Melles DC, de Jong MD, Schultsz C; COMBAT Consortium; Dantas G, Penders J. D'Souza AW, et al. Genome Med. 2021 Jun 7;13(1):79. doi: 10.1186/s13073-021-00893-z. Genome Med. 2021. PMID: 34092249 Free PMC article. - High fecal carriage of blaCTX-M, blaCMY-2, and plasmid-mediated quinolone resistance genes among healthy Korean people in a metagenomic analysis.
Kim J, Park KY, Park HK, Hwang HS, Seo MR, Kim B, Cho Y, Rho M, Pai H. Kim J, et al. Sci Rep. 2021 Mar 12;11(1):5874. doi: 10.1038/s41598-021-84974-4. Sci Rep. 2021. PMID: 33712656 Free PMC article. - Characterization of carbapenemases, extended spectrum β-lactamases, quinolone resistance and aminoglycoside resistance determinants in carbapenem-non-susceptible Escherichia coli from a teaching hospital in Chongqing, Southwest China.
Zhang C, Xu X, Pu S, Huang S, Sun J, Yang S, Zhang L. Zhang C, et al. Infect Genet Evol. 2014 Oct;27:271-6. doi: 10.1016/j.meegid.2014.07.031. Epub 2014 Aug 10. Infect Genet Evol. 2014. PMID: 25107431 - Molecular characteristics of extended-spectrum β-lactamases in clinical isolates of Enterobacteriaceae from the Philippines.
Kanamori H, Navarro RB, Yano H, Sombrero LT, Capeding MR, Lupisan SP, Olveda RM, Arai K, Kunishima H, Hirakata Y, Kaku M. Kanamori H, et al. Acta Trop. 2011 Oct-Nov;120(1-2):140-5. doi: 10.1016/j.actatropica.2011.07.007. Epub 2011 Jul 28. Acta Trop. 2011. PMID: 21820398 - Emergence of plasmid-mediated resistance to quinolones in Enterobacteriaceae.
Nordmann P, Poirel L. Nordmann P, et al. J Antimicrob Chemother. 2005 Sep;56(3):463-9. doi: 10.1093/jac/dki245. Epub 2005 Jul 14. J Antimicrob Chemother. 2005. PMID: 16020539 Review.
Cited by
- Meta-analysis of proportion estimates of Extended-Spectrum-Beta-Lactamase-producing Enterobacteriaceae in East Africa hospitals.
Sonda T, Kumburu H, van Zwetselaar M, Alifrangis M, Lund O, Kibiki G, Aarestrup FM. Sonda T, et al. Antimicrob Resist Infect Control. 2016 May 14;5:18. doi: 10.1186/s13756-016-0117-4. eCollection 2016. Antimicrob Resist Infect Control. 2016. PMID: 27186369 Free PMC article. Review. - No nosocomial transmission under standard hygiene precautions in short term contact patients in case of an unexpected ESBL or Q&A E. coli positive patient: a one-year prospective cohort study within three regional hospitals.
Souverein D, Euser SM, Herpers BL, Hattink C, Houtman P, Popma A, Kluytmans J, Rossen JWA, Den Boer JW. Souverein D, et al. Antimicrob Resist Infect Control. 2017 Jun 26;6:69. doi: 10.1186/s13756-017-0228-6. eCollection 2017. Antimicrob Resist Infect Control. 2017. PMID: 28670449 Free PMC article. - Travel-Related Antimicrobial Resistance: A Systematic Review.
Bokhary H, Pangesti KNA, Rashid H, Abd El Ghany M, Hill-Cawthorne GA. Bokhary H, et al. Trop Med Infect Dis. 2021 Jan 16;6(1):11. doi: 10.3390/tropicalmed6010011. Trop Med Infect Dis. 2021. PMID: 33467065 Free PMC article. Review. - Investigating the burden of antibiotic resistance in ethnic minority groups in high-income countries: protocol for a systematic review and meta-analysis.
Lishman H, Aylin P, Alividza V, Castro-Sanchez E, Chatterjee A, Mariano V, Johnson AP, Jeraj S, Costelloe C. Lishman H, et al. Syst Rev. 2017 Dec 11;6(1):251. doi: 10.1186/s13643-017-0654-9. Syst Rev. 2017. PMID: 29228985 Free PMC article. - Association of knowledge and beliefs with the misuse of antibiotics in parents: A study in Beirut (Lebanon).
Mallah N, Badro DA, Figueiras A, Takkouche B. Mallah N, et al. PLoS One. 2020 Jul 22;15(7):e0232464. doi: 10.1371/journal.pone.0232464. eCollection 2020. PLoS One. 2020. PMID: 32697808 Free PMC article.
References
- Wright GD. The antibiotic resistome. Expert Opinion on Drug Discovery. 2010;5:779–88. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources