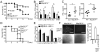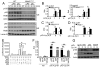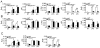Noncanonical NF-κB pathway controls the production of type I interferons in antiviral innate immunity - PubMed (original) (raw)
Noncanonical NF-κB pathway controls the production of type I interferons in antiviral innate immunity
Jin Jin et al. Immunity. 2014.
Abstract
Production of type I interferons (IFN-I) is a crucial innate immune mechanism against viral infections. IFN-I induction is subject to negative regulation by both viral and cellular factors, but the underlying mechanism remains unclear. We report that the noncanonical NF-κB pathway was stimulated along with innate immune cell differentiation and viral infections and had a vital role in negatively regulating IFN-I induction. Genetic deficiencies in major components of the noncanonical NF-κB pathway caused IFN-I hyperinduction and rendered cells and mice substantially more resistant to viral infection. Noncanonical NF-κB suppressed signal-induced histone modifications at the Ifnb promoter, an action that involved attenuated recruitment of the transcription factor RelA and a histone demethylase, JMJD2A. These findings reveal an unexpected function of the noncanonical NF-κB pathway and highlight an important mechanism regulating antiviral innate immunity.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures
Figure 1. NIK deficiency potentiates antiviral immunity
(A–C) Age-matched (6–8 weeks old) WT and _Map3k14_−/− mice, bred to the _Rag1_−/− background, were infected i.v. with VSV (2×107 PFU per mouse). (A) Survival curves (n=12). (B) Tissue VSV titers on day 3 of infection. (C) Serum concentrations of IFN-α and IFN-β at 12 h of infection. Data are presented as mean ± S.D. of six animals. (D–E) WT and _Map3k14_−/− mice, bred to the _Rag1_−/−-_Ifnar1_−/− background, were infected i.v. with VSV (2×107 PFU per mouse). (D) Survival curves (n=6). (E) Tissue VSV titers on day 3 of infection. Data are presented as mean ± S.D. of six animals. (F) WT or _Map3k14_−/− primary MEFs were infected with GFP-expressing VSV (VSV-GFP) at a MOI of 0.1 for 24 hr. Data are presented as a representative picture, showing the infected (GFP+) and total (bright field) cells (left). The sacle bar indicated as length of 1000μm. The summary graph of flow cytometric quantification of the infected cells (right). Data are representative of 3 independent experiments, and statistical analyses represent variations in technical replicates. *P<0.05. See also Figure S1.
Figure 2. NIK negatively regulates IFN-I induction
(A and B) WT or _Map3k14_−/− primary MEFs were infected with VSV (A) or SeV (B). The relative mRNA amount and protein concentration were determined by QPCR and ELISA, respectively. Data are presented as mean ± S.D. (C) QPCR analysis of relative Ifna and Ifnb mRNA amounts in WT and _Map3k14_−/− MEF cells stimulated with Lipofectamine-transfected poly(I:C) (pIC) for the indicated time periods. (D–F) BMDMs derived from WT or _Map3k14_−/− mice were infected with SeV (D) or stimulated with LPS and poly(I:C) (E and F). Relative mRNA amounts (D and E) and protein concentration (F) of IFNβ were determined by QPCR and ELISA, respectively. (G) QPCR analysis of Ifnb mRNA expression in WT and NIKΔT3 BMDMs stimulated with LPS or poly(I:C). (H) QPCR analysis of Ifna and Ifnb mRNA expression in FLT3L-differentiated pDC (F-pDC) and cDC (F-cDC) cells and GM-CSF-differentiated cDC (G-cDC) cells stimulated as indicated. All QPCR data are presented as fold relative to the amount of an internal control, Actb. Data are representative of three-four independent experiments, and statistical analyses represent variations in technical replicates. *P<0.05 and **P<0.01. See also Figure S1.
Figure 3. NIK-dependent noncanonical NF-κB signaling negatively regulates IFN-I induction
(A) Immunoblot analysis of the indicated phosphorylated (P-) and total proteins in the cytoplasmic (CE) and nuclear (NE) extracts of WT and _Map3k14_−/− BMDMs stimulated with LPS.(B and C) Immunoblot analysis of the indicated proteins in the cytoplasmic (CE) and nuclear (NE) extracts of WT and _Map3k14_−/− MEFs infected with VSV or SeV (B) or stimulated with Lipofectamine-transfected poly(I:C) (C). (D and E) Immunoblot analysis of the indicated proteins in the cytoplasmic (CE) and nuclear (NE) extracts of _Map3k14_−/− (D), _Nfkb2_lym1/+ (E) or their WT control BMDMs stimulated with LPS and poly(I:C) as indicated. (F) QPCR analysis of Ifnb mRNA amounts (fold relative to the internal control Actb mRNA) of WT and _Nfkb2_lym1/+BMDMs stimulated with LPS and poly(I:C). *P<0.05 and **P<0.01. Data in all panels are representative of three independent experiments, and statistical analyses represent variations in technical replicates. *P<0.05 and **P<0.01. See also Figures S2 and S3.
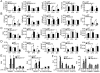
Figure 4. M-CSF induces noncanonical NF-κB signaling
(A and B) Immunoblot analysis of the indicated proteins in the cytoplasmic (CE) and nuclear (NE) extracts of bone marrow (BM) cells or M-CSF-induced BMDMs (BMM) from WT and _Map3k14_−/− mice. (C) Immunoblot analysis of the indicated proteins in whole-cell lysates of WT or _Map3k14_−/− bone marrow cells stimulated with M-CSF for the indicated time points. (D) Bone marrow cells were stimulated with M-CSF in the presence of the proteasome inhibitor MG132. TRAF2 and TRAF3 were immunoprecipitated from denatured cell lysates, and ubiquitinated TRAF2 and TRAF3 was detected by immunoblot using an antibody detecting K48-linked polyubiquitin chains. (E) WT bone marrow cells were stimulated with M-CSF in the presence of the proteasome inhibitor MG132. Whole-cell lysates were subjected to M-CSFR IP followed by detecting MCSFR-associated TRAF2 and TRAF3 by IB (top two panels). Cell lysates were also directly subjected to direct IB (lower four panels). (F) Immunoblot analysis of the indicated proteins in the cytoplasmic (CE) and nuclear (NE) extracts of FLT3L or GM-CSF generated dendritc cells from WT and _Map3k14_−/− mice. (G) D2SC/1 dendritic cell line constituted with FLT3 (D2SC/1-FLT3) was stimulated with GM-CSF or FLT3L as indicated and subjected to immunoblot analyses. Data in all panels are representative of two-three independent experiments. See also Figure S4.
Figure 5. Noncanonical NF-κB members suppress IFN-I induction
(A) Immunoblot analysis of the indicated proteins in the cytoplasmic (CE) and nuclear (NE) extracts of WT or _Nfkb2_xdr/xdr BMDMs stimulated with LPS and poly(I:C). (B) QPCR analysis of relative amounts of Ifnb mRNAs (fold of the internal control Actb mRNA) in the WT and _Nfkb2_xdr/xdr BMDMs stimulated with LPS and poly(I:C) (pIC). (C and D) QPCR analysis of relative amounts of Ifnb mRNA in xdr/xdr and WT control MEFs (C) or _Relb_−/− and WT control MEFs (D) stimulated with Lipofectamine-transfected poly(I:C). (E) HEK293 cells were transfected with an _Ifnb_-luciferase reporter plasmid in the presence (+) or absence (−) of the indicated empty vector or expression plasmids. Luciferase assays were performed as fold based on empty vector group 36 h after transfection. (F and G) _Nfkb2_xdr/xdr MEFs were reconstituted with GFP, p100, p52 and p100SSAA. The cells were either not treated (NT) or stimulated with Lipofectamine-transfected poly(I:C) and subjected QPCR (F) or IB (G) analysis. Data in all panels are representative of 2–3 independent experiments, and statistical analyses represent variations in technical replicates. *P<0.05 and **P<0.01.
Figure 6. RelA physically interacts with JMJD2A and binds to Ifnb promoter in a manner that is suppressed by noncanonical NF-κB
(A–C) BMDMs prepared from _Map3k14_−/− (A), NIKΔT3 transgenic (B), or _Nfkb2_xdr/xdr (C) mice and their WT littermate controls were stimulated for 1 h with LPS or poly(I:C). ChIP assays were performed and quantified by QPCR to detect the binding of NF-κB family members to the Ifnb promoter. Data are presented as percentage of the total input DNA. Data are representative of two-three independent experiments, and statistical analyses represent variations in technical replicates. *P<0.05 and **P<0.01. See also Figure S5.
Figure 7. NIK regulates histone modifications and JMJD2A binding at the Ifnb promoter
(A–D) BMDMs derived from _Map3k14_−/−, NIKΔT3 transgenic, or their WT control mice were stimulated with LPS or poly(I:C) for 1h. ChIP assays were performed to detect histone modifications (A and B) and binding of the indicated factors (C and D) at the Ifnb promoter. The Y axis is percentage (%) based on total H3 for A–B, and percentage (%) based on total input DNA for C–D. Data are representative of three independent experiments, and statistical analyses represent variations in technical replicates. (E) WT and _Map3k14_−/− BMDMs were pretreated for 12 h with 20 mM of a JMJD2 inhibitor (5-carboxy-8HQ) and then stimulated with LPS or poly(I:C) as indicated. The relative amount of Ifnb mRNAs were quantified by QPCR and presented as fold relative to the internal Actb mRNA control. (F) WT and _Map3k14_−/− BMDMs were infected with two different JMJD2A shRNAs or a non-silencing control shRNA and then stimulated with LPS and poly(I:C) as indicated. The relative amount of Ifnb mRNAs were quantified by QPCR and presented as fold relative to the internal Actb mRNA control. Data in all panels are representative of two-three independent experiments, and statistical analyses represent variations in technical replicates. *P<0.05 and **P<0.01. See also Figures S6 and S7
Similar articles
- IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway.
Fitzgerald KA, McWhirter SM, Faia KL, Rowe DC, Latz E, Golenbock DT, Coyle AJ, Liao SM, Maniatis T. Fitzgerald KA, et al. Nat Immunol. 2003 May;4(5):491-6. doi: 10.1038/ni921. Nat Immunol. 2003. PMID: 12692549 - SAMHD1 suppresses innate immune responses to viral infections and inflammatory stimuli by inhibiting the NF-κB and interferon pathways.
Chen S, Bonifati S, Qin Z, St Gelais C, Kodigepalli KM, Barrett BS, Kim SH, Antonucci JM, Ladner KJ, Buzovetsky O, Knecht KM, Xiong Y, Yount JS, Guttridge DC, Santiago ML, Wu L. Chen S, et al. Proc Natl Acad Sci U S A. 2018 Apr 17;115(16):E3798-E3807. doi: 10.1073/pnas.1801213115. Epub 2018 Apr 2. Proc Natl Acad Sci U S A. 2018. PMID: 29610295 Free PMC article. - Zinc finger protein 64 promotes Toll-like receptor-triggered proinflammatory and type I interferon production in macrophages by enhancing p65 subunit activation.
Wang C, Liu X, Liu Y, Zhang Q, Yao Z, Huang B, Zhang P, Li N, Cao X. Wang C, et al. J Biol Chem. 2013 Aug 23;288(34):24600-8. doi: 10.1074/jbc.M113.473397. Epub 2013 Jul 15. J Biol Chem. 2013. PMID: 23857586 Free PMC article. Retracted. - Intrinsic cellular defenses against virus infection by antiviral type I interferon.
Boo KH, Yang JS. Boo KH, et al. Yonsei Med J. 2010 Jan;51(1):9-17. doi: 10.3349/ymj.2010.51.1.9. Epub 2009 Dec 29. Yonsei Med J. 2010. PMID: 20046508 Free PMC article. Review. - Induction and regulation of IFNs during viral infections.
Malmgaard L. Malmgaard L. J Interferon Cytokine Res. 2004 Aug;24(8):439-54. doi: 10.1089/1079990041689665. J Interferon Cytokine Res. 2004. PMID: 15320958 Review.
Cited by
- Proteomics Analysis of Duck Lung Tissues in Response to Highly Pathogenic Avian Influenza Virus.
Vijayakumar P, Mishra A, Deka RP, Pinto SM, Subbannayya Y, Sood R, Prasad TSK, Raut AA. Vijayakumar P, et al. Microorganisms. 2024 Jun 25;12(7):1288. doi: 10.3390/microorganisms12071288. Microorganisms. 2024. PMID: 39065055 Free PMC article. - Non-canonical NF-κB Antagonizes STING Sensor-Mediated DNA Sensing in Radiotherapy.
Hou Y, Liang H, Rao E, Zheng W, Huang X, Deng L, Zhang Y, Yu X, Xu M, Mauceri H, Arina A, Weichselbaum RR, Fu YX. Hou Y, et al. Immunity. 2018 Sep 18;49(3):490-503.e4. doi: 10.1016/j.immuni.2018.07.008. Epub 2018 Aug 28. Immunity. 2018. PMID: 30170810 Free PMC article. - Glucocorticoids, their uses, sexual dimorphisms, and diseases: new concepts, mechanisms, and discoveries.
Martinez GJ, Appleton M, Kipp ZA, Loria AS, Min B, Hinds TD Jr. Martinez GJ, et al. Physiol Rev. 2024 Jan 1;104(1):473-532. doi: 10.1152/physrev.00021.2023. Epub 2023 Sep 21. Physiol Rev. 2024. PMID: 37732829 Free PMC article. Review. - Miz1 represses type I interferon production and limits viral clearance during influenza A virus infection.
Wu W, Arunagiri V, Do-Umehara HC, Chen C, Gu S, Biswas I, Ridge KM, Budinger GRS, Liu S, Liu J. Wu W, et al. Sci Signal. 2024 Apr 9;17(831):eadg7867. doi: 10.1126/scisignal.adg7867. Epub 2024 Apr 9. Sci Signal. 2024. PMID: 38593156 Free PMC article. - JNK1 Derived from Orange-Spotted Grouper, Epinephelus coioides, Involving in the Evasion and Infection of Singapore Grouper Iridovirus (SGIV).
Guo M, Wei J, Huang X, Zhou Y, Yan Y, Qin Q. Guo M, et al. Front Microbiol. 2016 Feb 10;7:121. doi: 10.3389/fmicb.2016.00121. eCollection 2016. Front Microbiol. 2016. PMID: 26903999 Free PMC article.
References
- Asano M, Hayashi M, Yoshida E, Kawade Y, Iwakura Y. Induction of interferon-alpha by interferon-beta, but not of interferon-beta by interferon-alpha, in the mouse. Virology. 1990;176:30–38. - PubMed
- Berger SL. Histone modifications in transcriptional regulation. Curr. Opin. Genet. Dev. 2002;12:142–148. - PubMed
- Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat. Rev. Genet. 2009;10:295–304. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI064639/AI/NIAID NIH HHS/United States
- P30CA016672/CA/NCI NIH HHS/United States
- AI064639/AI/NIAID NIH HHS/United States
- R01 AI057555/AI/NIAID NIH HHS/United States
- GM84459/GM/NIGMS NIH HHS/United States
- R37 AI064639/AI/NIAID NIH HHS/United States
- R01 AI104519/AI/NIAID NIH HHS/United States
- P30 CA016672/CA/NCI NIH HHS/United States
- AI104519/AI/NIAID NIH HHS/United States
- AI057555/AI/NIAID NIH HHS/United States
- R01 GM084459/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases